Herpesviruses and chromosomal integration
- PMID: 20844040
- PMCID: PMC2976420
- DOI: 10.1128/JVI.01169-10
Herpesviruses and chromosomal integration
Abstract
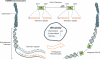
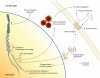
Herpesviruses are members of a diverse family of viruses that colonize all vertebrates from fish to mammals. Although more than one hundred herpesviruses exist, all are nearly identical architecturally, with a genome consisting of a linear double-stranded DNA molecule (100 to 225 kbp) protected by an icosahedral capsid made up of 162 hollow-centered capsomeres, a tegument surrounding the nucleocapsid, and a viral envelope derived from host membranes. Upon infection, the linear viral DNA is delivered to the nucleus, where it circularizes to form the viral episome. Depending on several factors, the viral cycle can proceed either to a productive infection or to a state of latency. In either case, the viral genetic information is maintained as extrachromosomal circular DNA. Interestingly, however, certain oncogenic herpesviruses such as Marek's disease virus and Epstein-Barr virus can be found integrated at low frequencies in the host's chromosomes. These findings have mostly been viewed as anecdotal and considered exceptions rather than properties of herpesviruses. In recent years, the consistent and rather frequent detection (in approximately 1% of the human population) of human herpesvirus 6 (HHV-6) viral DNA integrated into human chromosomes has spurred renewed interest in our understanding of how these viruses infect, replicate, and propagate themselves. In this review, we provide a historical perspective on chromosomal integration by herpesviruses and present the current state of knowledge on integration by HHV-6 with the possible clinical implications associated with viral integration.
Figures

References
-
- Ablashi, D. V., N. Balachandran, S. F. Josephs, C. L. Hung, G. R. Krueger, B. Kramarsky, S. Z. Salahuddin, and R. C. Gallo. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184:545-552. - PubMed
-
- Achour, A., I. Malet, F. Le Gal, A. Dehee, A. Gautheret-Dejean, P. Bonnafous, and H. Agut. 2008. Variability of gB and gH genes of human herpesvirus-6 among clinical specimens. J. Med. Virol. 80:1211-1221. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

