Phylogeny and identification of Nocardia species on the basis of multilocus sequence analysis
- PMID: 20844218
- PMCID: PMC3008441
- DOI: 10.1128/JCM.00883-10
Phylogeny and identification of Nocardia species on the basis of multilocus sequence analysis
Erratum in
-
Erratum for McTaggart et al., "Phylogeny and Identification of Nocardia Species on the Basis of Multilocus Sequence Analysis".J Clin Microbiol. 2020 Jul 23;58(8):e00961-20. doi: 10.1128/JCM.00961-20. Print 2020 Jul 23. J Clin Microbiol. 2020. PMID: 32703887 Free PMC article. No abstract available.
Abstract
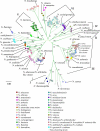
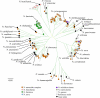
Nocardia species identification is difficult due to a complex and rapidly changing taxonomy, the failure of 16S rRNA and cellular fatty acid analysis to discriminate many species, and the unreliability of biochemical testing. Here, Nocardia species identification was achieved through multilocus sequence analysis (MLSA) of gyrase B of the β subunit of DNA topoisomerase (gyrB), 16S rRNA (16S), subunit A of SecA preprotein translocase (secA1), the 65-kDa heat shock protein (hsp65), and RNA polymerase (rpoB) applied to 190 clinical, 36 type, and 11 reference strains. Phylogenetic analysis resolved 30 sequence clusters with high (>85%) bootstrap support. Since most clusters contained a single type strain and the analysis corroborated current knowledge of Nocardia taxonomy, the sequence clusters were equated with species clusters and MLSA was deemed appropriate for species identification. By comparison, single-locus analysis was inadequate because it failed to resolve species clusters, partly due to the presence of foreign alleles in 22.1% of isolates. While MLSA identified the species of the majority (71.3%) of strains, it also identified clusters that may correspond to new species. The correlation of the identities by MLSA with those determined on the basis of microscopic examination, biochemical testing, and fatty acid analysis was 95%; however, MLSA was more discriminatory. Nocardia cyriacigeorgica (21.58%) and N. farcinica (14.74%) were the most frequently encountered species among clinical isolates. In summary, five-locus MLSA is a reliable method of elucidating taxonomic data to inform Nocardia species identification; however, three-locus (gyrB-16S-secA1) or four-locus (gyrB-16S-secA1-hsp65) MLSA was nearly as reliable, correctly identifying 98.5% and 99.5% of isolates, respectively, and would be more feasible for routine use in a clinical reference microbiology laboratory.
Figures
Similar articles
-
Accurate Identification of Common Pathogenic Nocardia Species: Evaluation of a Multilocus Sequence Analysis Platform and Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry.PLoS One. 2016 Jan 25;11(1):e0147487. doi: 10.1371/journal.pone.0147487. eCollection 2016. PLoS One. 2016. PMID: 26808813 Free PMC article.
-
Molecular identification and phylogenetic relationships of clinical Nocardia isolates.Antonie Van Leeuwenhoek. 2019 Dec;112(12):1755-1766. doi: 10.1007/s10482-019-01296-2. Epub 2019 Jul 26. Antonie Van Leeuwenhoek. 2019. PMID: 31350617
-
secA1 gene sequence polymorphisms for species identification of Nocardia species and recognition of intraspecies genetic diversity.J Clin Microbiol. 2010 Nov;48(11):3928-34. doi: 10.1128/JCM.01113-10. Epub 2010 Sep 1. J Clin Microbiol. 2010. PMID: 20810768 Free PMC article.
-
Comparison of restriction enzyme pattern analysis and full gene sequencing of 16S rRNA gene for Nocardia species identification, the first report of Nocardia transvalensis isolated of sputum from Iran, and review of the literature.Antonie Van Leeuwenhoek. 2016 Oct;109(10):1285-98. doi: 10.1007/s10482-016-0746-x. Epub 2016 Sep 9. Antonie Van Leeuwenhoek. 2016. PMID: 27613736 Review.
-
Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy.Clin Microbiol Rev. 2006 Apr;19(2):259-82. doi: 10.1128/CMR.19.2.259-282.2006. Clin Microbiol Rev. 2006. PMID: 16614249 Free PMC article. Review.
Cited by
-
Identification, typing, and phylogenetic relationships of the main clinical Nocardia species in spain according to their gyrB and rpoB genes.J Clin Microbiol. 2013 Nov;51(11):3602-8. doi: 10.1128/JCM.00515-13. Epub 2013 Aug 21. J Clin Microbiol. 2013. PMID: 23966490 Free PMC article.
-
Molecular identification and antibiotic resistance pattern of actinomycetes isolates among immunocompromised patients in Iran, emerging of new infections.Sci Rep. 2021 May 24;11(1):10745. doi: 10.1038/s41598-021-90269-5. Sci Rep. 2021. PMID: 34031507 Free PMC article.
-
Epidemiology of Nocardia Species at a Tertiary Hospital in Southern Taiwan, 2012 to 2020: MLSA Phylogeny and Antimicrobial Susceptibility.Antibiotics (Basel). 2022 Oct 19;11(10):1438. doi: 10.3390/antibiotics11101438. Antibiotics (Basel). 2022. PMID: 36290097 Free PMC article.
-
Genotyping of Nocardia farcinica with multilocus sequence typing.Eur J Clin Microbiol Infect Dis. 2016 May;35(5):771-8. doi: 10.1007/s10096-016-2596-x. Epub 2016 Mar 14. Eur J Clin Microbiol Infect Dis. 2016. PMID: 26972429
-
Accurate Identification of Common Pathogenic Nocardia Species: Evaluation of a Multilocus Sequence Analysis Platform and Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry.PLoS One. 2016 Jan 25;11(1):e0147487. doi: 10.1371/journal.pone.0147487. eCollection 2016. PLoS One. 2016. PMID: 26808813 Free PMC article.
References
-
- Adekambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. - PubMed
-
- Agterof, M. J., T. van der Bruggen, M. Tersmette, E. J. ter Borg, J. M. M. van den Bosch, and D. H. Biesma. 2007. Nocardiosis: a case series and a mini review of clinical and microbiological features. Neth. J. Med. 65:199-202. - PubMed
-
- Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. - PubMed
-
- Brady, C., I. Cleenwerck, S. Venter, M. Vancanneyt, J. Swings, and T. Coutinho. 2008. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 31:447-460. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

