Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo
- PMID: 20844538
- PMCID: PMC3773546
- DOI: 10.1038/nature09356
Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo
Abstract
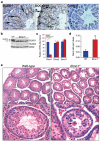
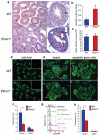
Apoptosis and the subsequent clearance of dying cells occurs throughout development and adult life in many tissues. Failure to promptly clear apoptotic cells has been linked to many diseases. ELMO1 is an evolutionarily conserved cytoplasmic engulfment protein that functions downstream of the phosphatidylserine receptor BAI1, and, along with DOCK1 and the GTPase RAC1, promotes internalization of the dying cells. Here we report the generation of ELMO1-deficient mice, which we found to be unexpectedly viable and grossly normal. However, they had a striking testicular pathology, with disrupted seminiferous epithelium, multinucleated giant cells, uncleared apoptotic germ cells and decreased sperm output. Subsequent in vitro and in vivo analyses revealed a crucial role for ELMO1 in the phagocytic clearance of apoptotic germ cells by Sertoli cells lining the seminiferous epithelium. The engulfment receptor BAI1 and RAC1 (upstream and downstream of ELMO1, respectively) were also important for Sertoli-cell-mediated engulfment. Collectively, these findings uncover a selective requirement for ELMO1 in Sertoli-cell-mediated removal of apoptotic germ cells and make a compelling case for a relationship between engulfment and tissue homeostasis in vivo.
Figures


References
-
- Henson PM. Dampening inflammation. Nat Immunol. 2005;6:1179–1181. - PubMed
-
- Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 140:619–630. - PubMed
-
- Gregory CD, Pound JD. Results of Defective Clearance of Apoptotic Cells: Lessons from Knock-out Mouse Models. In: Krysko DV, Vandenabeele P, editors. Phagocytosis of Dying Cells. Vol. 1. Springer Science; 2009. pp. 271–298.
-
- Zhou Z, Caron E, Hartwieg E, Hall A, Horvitz HR. The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev Cell. 2001;1:477–489. - PubMed
-
- Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NY. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev Cell. 2001;1:491–502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

