The CC-NB-LRR-type Rdg2a resistance gene confers immunity to the seed-borne barley leaf stripe pathogen in the absence of hypersensitive cell death
- PMID: 20844752
- PMCID: PMC2937021
- DOI: 10.1371/journal.pone.0012599
The CC-NB-LRR-type Rdg2a resistance gene confers immunity to the seed-borne barley leaf stripe pathogen in the absence of hypersensitive cell death
Abstract
Background: Leaf stripe disease on barley (Hordeum vulgare) is caused by the seed-transmitted hemi-biotrophic fungus Pyrenophora graminea. Race-specific resistance to leaf stripe is controlled by two known Rdg (Resistance to Drechslera graminea) genes: the H. spontaneum-derived Rdg1a and Rdg2a, identified in H. vulgare. The aim of the present work was to isolate the Rdg2a leaf stripe resistance gene, to characterize the Rdg2a locus organization and evolution and to elucidate the histological bases of Rdg2a-based leaf stripe resistance.
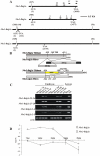
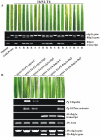
Principal findings: We describe here the positional cloning and functional characterization of the leaf stripe resistance gene Rdg2a. At the Rdg2a locus, three sequence-related coiled-coil, nucleotide-binding site, and leucine-rich repeat (CC-NB-LRR) encoding genes were identified. Sequence comparisons suggested that paralogs of this resistance locus evolved through recent gene duplication, and were subjected to frequent sequence exchange. Transformation of the leaf stripe susceptible cv. Golden Promise with two Rdg2a-candidates under the control of their native 5' regulatory sequences identified a member of the CC-NB-LRR gene family that conferred resistance against the Dg2 leaf stripe isolate, against which the Rdg2a-gene is effective. Histological analysis demonstrated that Rdg2a-mediated leaf stripe resistance involves autofluorescing cells and prevents pathogen colonization in the embryos without any detectable hypersensitive cell death response, supporting a cell wall reinforcement-based resistance mechanism.
Conclusions: This work reports about the cloning of a resistance gene effective against a seed borne disease. We observed that Rdg2a was subjected to diversifying selection which is consistent with a model in which the R gene co-evolves with a pathogen effector(s) gene. We propose that inducible responses giving rise to physical and chemical barriers to infection in the cell walls and intercellular spaces of the barley embryo tissues represent mechanisms by which the CC-NB-LRR-encoding Rdg2a gene mediates resistance to leaf stripe in the absence of hypersensitive cell death.
Conflict of interest statement
Figures





References
-
- Delogu G, Porta-Puglia A, Stanca AM, Vannacci G. Interaction between barley and Pyrenophora graminea: an overview of research in Italy. Rachis. 1995;14:29–34.
-
- Mueller KJ, Valè G, Enneking D. Selection of resistant spring barley accessions after natural infection with leaf stripe (Pyrenophora graminea) under organic farming conditions in Germany and by sandwich test. J Plant Pathol. 2003;85:9–14.
-
- Platenkamp R. Investigations on the infections pathway of Drechslera graminea in germinating barley. Royal Veterinary and Agricultural University, Yearbook. 1976:49–64.
-
- Hammouda AM. Variability of Drechslera graminea, the causal fungus of leaf stripe of barley. Acta Phytopathol Entomol Hung. 1988;23:73–80.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

