Evaluation of phenoxybenzamine in the CFA model of pain following gene expression studies and connectivity mapping
- PMID: 20846436
- PMCID: PMC2949723
- DOI: 10.1186/1744-8069-6-56
Evaluation of phenoxybenzamine in the CFA model of pain following gene expression studies and connectivity mapping
Abstract
Background: We have previously used the rat 4 day Complete Freund's Adjuvant (CFA) model to screen compounds with potential to reduce osteoarthritic pain. The aim of this study was to identify genes altered in this model of osteoarthritic pain and use this information to infer analgesic potential of compounds based on their own gene expression profiles using the Connectivity Map approach.
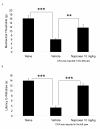
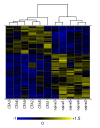
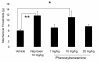
Results: Using microarrays, we identified differentially expressed genes in L4 and L5 dorsal root ganglia (DRG) from rats that had received intraplantar CFA for 4 days compared to matched, untreated control animals. Analysis of these data indicated that the two groups were distinguishable by differences in genes important in immune responses, nerve growth and regeneration. This list of differentially expressed genes defined a "CFA signature". We used the Connectivity Map approach to identify pharmacologic agents in the Broad Institute Build02 database that had gene expression signatures that were inversely related ('negatively connected') with our CFA signature. To test the predictive nature of the Connectivity Map methodology, we tested phenoxybenzamine (an alpha adrenergic receptor antagonist) - one of the most negatively connected compounds identified in this database - for analgesic activity in the CFA model. Our results indicate that at 10 mg/kg, phenoxybenzamine demonstrated analgesia comparable to that of Naproxen in this model.
Conclusion: Evaluation of phenoxybenzamine-induced analgesia in the current study lends support to the utility of the Connectivity Map approach for identifying compounds with analgesic properties in the CFA model.
Figures

References
-
- Costigan M, Befort K, Karchewski L, Griffin R, D'Urso D, Allchorne A, Sitarski J, Mannion J, Pratt R, Woolf C. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neuroscience. 2002;3:16. doi: 10.1186/1471-2202-3-16. - DOI - PMC - PubMed
-
- Rodriguez Parkitna JKM, Kaminska-Chowaniec D, Obara I, Mika J, Przewlocka B, Przewlocki R. Comparison of gene expression profiles in neuropathic and inflammatory pain. J Physiol Pharmacol. 2006;57:14. - PubMed
-
- Rabert D, Xiao Y, Yiangou Y, Kreder D, Sangameswaran L, Segal MR, Hunt CA, Birch R, Anand P. Plasticity of gene expression in injured human dorsal root ganglia revealed by GeneChip oligonucleotide microarrays. Journal of Clinical Neuroscience. 2004;11:289–299. doi: 10.1016/j.jocn.2003.05.008. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

