Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8
- PMID: 20846456
- PMCID: PMC2946291
- DOI: 10.1186/1476-4598-9-249
Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8
Abstract
Background: Colorectal cancer (CRC) has long been associated with bacteremia and/or endocarditis by Streptococcus gallolyticus member bacteria (SGMB) but the direct colonization of SGMB along with its molecular carcinogenic role, if any, has not been investigated. We assessed the colonization of SGMB in CRC patients with history of bacteremia (CRC-w/bac) and without history of bacteremia (CRC-wo/bac) by isolating SGMB from feces, mucosal surfaces of colorectum, and colorectal tissues and detecting SGMB DNA, via PCR and in situ hybridization (ISH) assays targeting SodA gene in colorectal tissues. Moreover, mRNA of IL1, IL-8, COX-2, IFN-γ, c-Myc, and Bcl-2 in colorectal tissues of studied groups was assessed via ISH and RT-PCR.
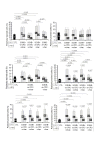
Results: SGMB were found to be remarkably isolated in tumorous (TU) and non-tumorous (NTU) tissues of CRC-w/bac, 20.5% and 17.3%, and CRC-wo/bac, 12.8% and 11.5%, respectively while only 2% of control tissues revealed SGMB (P < 0.05); such contrast was not found in mucosal and fecal isolation of SGMB. The positive detection of SGMB DNA in TU and NTU of CRC-w/bac and CRC-wo/bac via PCR, 48.7%, 35.9%, 32.7%, and 23%, respectively, and ISH, 46.1%, 30.7%, 28.8%, and 17.3%, respectively, was higher than in control tissues, 4 and 2%, respectively (P < 0.05). SGMB count measured via quantitative PCR of SGMB DNA in terms of copy number (CN), in TU and NTU of CRC-w/bac and CRC-wo/bac, 2.96-4.72, 1.29-2.81, 2.16-2.92, and 0.67-2.07 log10 CN/g respectively, showed higher colonization in TU than in NTU and in CRC-w/bac than in CRC-wo/bac (P < 0.05). The PCR-based mRNA ratio and ISH-based percentage of positively stained cells of IL-1, 1.77 and 70.3%, COX-2, 1.63 and 44.8%, and IL-8, 1.73 and 70.3%, respectively, rather than IFN-γ, c-Myc, and Bcl-2, were higher in SGMB positive patients than in control or SGMB negative patients (P < 0.05).
Conclusions: The current study indicated that colorectal cancer is remarkably associated with SGMB; moreover, molecular detection of SGMB in CRC was superior to link SGMB with CRC tumors highlighting a possible direct and active role of SGMB in CRC development through most probably inflammation-based sequel of tumor development or propagation via, but not limited to, IL-1, COX-2, and IL-8.
Figures





References
-
- Triantafillidis JK, Nasioulas G, Kosmidis PA. Colorectal cancer and inflammatory bowel disease: epidemiology, risk factors, mechanisms of carcinogenesis and prevention strategies. Anticancer Res. 2009;29(7):2727–2737. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

