Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures
- PMID: 20847124
- PMCID: PMC2962554
- DOI: 10.1093/gbe/evq056
Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures
Abstract
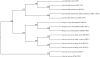
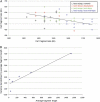
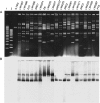
A large collection of Lactococcus lactis strains, including wild-type isolates and dairy starter cultures, were screened on the basis of their phenotype and the macrorestriction patterns produced from pulsed-field gel electrophoresis (PFGE) analysis of SmaI digests of genomic DNA. Three groups of dairy starter cultures, used for different purposes in the dairy industry, and a fourth group made up of strains isolated from the environment were selected for analysis of their chromosomal diversity using the endonuclease I-CeuI. Chromosome architecture was largely conserved with each strain having six copies of the rRNA genes, and the chromosome size of individual strains ranged between 2,240 and 2,688 kb. The origin of L. lactis strains showed the greatest correlation with chromosome size, and dairy strains, particularly those with the cremoris phenotype, had smaller chromosomes than wild-type strains. Overall, this study, coupled with analysis of the sequenced L. lactis genomes, provides evidence that defined strain dairy starter cultures have arisen from plant L. lactis strains. Adaptation of these strains to the dairy environment has involved loss of functions resulting in smaller chromosomes and acquisition of genes (usually plasmid associated) that facilitate growth in milk. We conclude that dairy starter cultures generally and the industrially used cremoris and diacetylactis phenotype strains in particular comprise a specialized group of L. lactis strains that have been selected to become an essential component of industrial processes and have evolved accordingly, so that they are no longer fit to survive outside the dairy environment.
Figures





References
-
- Beimfohr C, Ludwig W, Schleifer KH. Rapid genotypic differentiation of Lactococcus lactis subspecies and biovar. Syst Appl Microbiol. 1997;20:216–221.
-
- Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. - PubMed

