Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation
- PMID: 20847274
- PMCID: PMC2941909
- DOI: 10.1126/science.1191701
Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation
Abstract
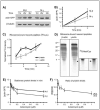
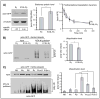
The mammalian cytoskeletal proteins β- and γ-actin are highly homologous, but only β-actin is amino-terminally arginylated in vivo, which regulates its function. We examined the metabolic fate of exogenously expressed arginylated and nonarginylated actin isoforms. Arginylated γ-actin, unlike β-, was highly unstable and was selectively ubiquitinated and degraded in vivo. This instability was regulated by the differences in the nucleotide coding sequence between the two actin isoforms, which conferred different translation rates. γ-actin was translated more slowly than β-actin, and this slower processing resulted in the exposure of a normally hidden lysine residue for ubiquitination, leading to the preferential degradation of γ-actin upon arginylation. This degradation mechanism, coupled to nucleotide coding sequence, may regulate protein arginylation in vivo.
Figures


Comment in
-
Cell biology. New roles for codon usage.Science. 2010 Sep 17;329(5998):1473-4. doi: 10.1126/science.1195567. Science. 2010. PMID: 20847254 No abstract available.
-
Biochemistry. Hidden code in the protein code.Nat Methods. 2010 Nov;7(11):874. doi: 10.1038/nmeth1110-874. Nat Methods. 2010. PMID: 21049579
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

