Ancient class of translocated oomycete effectors targets the host nucleus
- PMID: 20847293
- PMCID: PMC2951462
- DOI: 10.1073/pnas.1008491107
Ancient class of translocated oomycete effectors targets the host nucleus
Abstract
Pathogens use specialized secretion systems and targeting signals to translocate effector proteins inside host cells, a process that is essential for promoting disease and parasitism. However, the amino acid sequences that determine host delivery of eukaryotic pathogen effectors remain mostly unknown. The Crinkler (CRN) proteins of oomycete plant pathogens, such as the Irish potato famine organism Phytophthora infestans, are modular proteins with predicted secretion signals and conserved N-terminal sequence motifs. Here, we provide direct evidence that CRN N termini mediate protein transport into plant cells. CRN host translocation requires a conserved motif that is present in all examined plant pathogenic oomycetes, including the phylogenetically divergent species Aphanomyces euteiches that does not form haustoria, specialized infection structures that have been implicated previously in delivery of effectors. Several distinct CRN C termini localized to plant nuclei and, in the case of CRN8, required nuclear accumulation to induce plant cell death. These results reveal a large family of ubiquitous oomycete effector proteins that target the host nucleus. Oomycetes appear to have acquired the ability to translocate effector proteins inside plant cells relatively early in their evolution and before the emergence of haustoria. Finally, this work further implicates the host nucleus as an important cellular compartment where the fate of plant-microbe interactions is determined.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
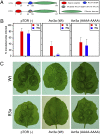
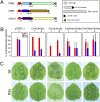

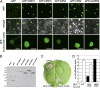
References
-
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. - PubMed
-
- Crabb BS, de Koning-Ward TF, Gilson PR. Protein export in Plasmodium parasites: From the endoplasmic reticulum to the vacuolar export machine. Int J Parasitol. 2010;40:509–513. - PubMed
-
- Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact. 2009;22:115–122. - PubMed
-
- Kamoun S. Groovy times: Filamentous pathogen effectors revealed. Curr Opin Plant Biol. 2007;10:358–365. - PubMed
-
- Haase S, de Koning-Ward TF. New insights into protein export in malaria parasites. Cell Microbiol. 2010;12:580–587. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

