Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea
- PMID: 20847933
- PMCID: PMC2933860
- DOI: 10.1155/2010/453642
Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea
Abstract
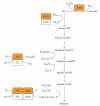
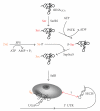
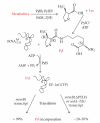
Methanogenic archaea are a group of strictly anaerobic microorganisms characterized by their strict dependence on the process of methanogenesis for energy conservation. Among the archaea, they are also the only known group synthesizing proteins containing selenocysteine or pyrrolysine. All but one of the known archaeal pyrrolysine-containing and all but two of the confirmed archaeal selenocysteine-containing protein are involved in methanogenesis. Synthesis of these proteins proceeds through suppression of translational stop codons but otherwise the two systems are fundamentally different. This paper highlights these differences and summarizes the recent developments in selenocysteine- and pyrrolysine-related research on archaea and aims to put this knowledge into the context of their unique energy metabolism.
Figures

References
-
- Kaji A, Kaji H, Novelli GD. A soluble amino acid incorporating system. Biochemical and Biophysical Research Communications. 1963;10(5):406–409. - PubMed
-
- Wold F. In vivo chemical modification of proteins (post-translational modification) Annual Review of Biochemistry. 1981;50:783–814. - PubMed
-
- Cone JE, Del Rio RM, Davis JN, Stadtman TC. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proceedings of the National Academy of Sciences of the United States of America. 1976;73(8):2659–2663. - PMC - PubMed
-
- Zinoni F, Birkmann A, Stadtman TC, Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli . Proceedings of the National Academy of Sciences of the United States of America. 1986;83(13):4650–4654. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

