Detection of clinically relevant exonic copy-number changes by array CGH
- PMID: 20848651
- PMCID: PMC3158569
- DOI: 10.1002/humu.21360
Detection of clinically relevant exonic copy-number changes by array CGH
Abstract
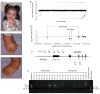
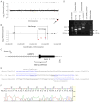
Array comparative genomic hybridization (aCGH) is a powerful tool for the molecular elucidation and diagnosis of disorders resulting from genomic copy-number variation (CNV). However, intragenic deletions or duplications--those including genomic intervals of a size smaller than a gene--have remained beyond the detection limit of most clinical aCGH analyses. Increasing array probe number improves genomic resolution, although higher cost may limit implementation, and enhanced detection of benign CNV can confound clinical interpretation. We designed an array with exonic coverage of selected disease and candidate genes and used it clinically to identify losses or gains throughout the genome involving at least one exon and as small as several hundred base pairs in size. In some patients, the detected copy-number change occurs within a gene known to be causative of the observed clinical phenotype, demonstrating the ability of this array to detect clinically relevant CNVs with subkilobase resolution. In summary, we demonstrate the utility of a custom-designed, exon-targeted oligonucleotide array to detect intragenic copy-number changes in patients with various clinical phenotypes.
© 2010 Wiley-Liss, Inc.
Figures




References
-
- Bartnik M, Tsai AC-H, Xia Z, Cheung SW, Stankiewicz P. Disruption of the SCN2A and SCN3A genes in a patient with mental retardation, neurobehavioral and psychiatric abnormalities, and a history of infantile seizures. Clin Genet. (in press) - PubMed
-
- Ben-Shachar S, Khajavi M, Withers MA, Shaw CA, van Bokhoven H, Brunner HG, Lupski JR. Dominant versus recessive traits conveyed by allelic mutations—to what extent is nonsense-mediated decay involved? Clin Genet. 2009;75:394–400. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

