Data-driven assessment of eQTL mapping methods
- PMID: 20849587
- PMCID: PMC2996998
- DOI: 10.1186/1471-2164-11-502
Data-driven assessment of eQTL mapping methods
Abstract
Background: The analysis of expression quantitative trait loci (eQTL) is a potentially powerful way to detect transcriptional regulatory relationships at the genomic scale. However, eQTL data sets often go underexploited because legacy QTL methods are used to map the relationship between the expression trait and genotype. Often these methods are inappropriate for complex traits such as gene expression, particularly in the case of epistasis.
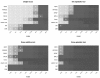
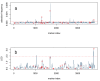
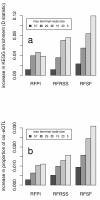
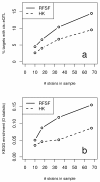
Results: Here we compare legacy QTL mapping methods with several modern multi-locus methods and evaluate their ability to produce eQTL that agree with independent external data in a systematic way. We found that the modern multi-locus methods (Random Forests, sparse partial least squares, lasso, and elastic net) clearly outperformed the legacy QTL methods (Haley-Knott regression and composite interval mapping) in terms of biological relevance of the mapped eQTL. In particular, we found that our new approach, based on Random Forests, showed superior performance among the multi-locus methods.
Conclusions: Benchmarks based on the recapitulation of experimental findings provide valuable insight when selecting the appropriate eQTL mapping method. Our battery of tests suggests that Random Forests map eQTL that are more likely to be validated by independent data, when compared to competing multi-locus and legacy eQTL mapping methods.
Figures
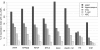

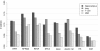



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

