Influenza virus M2 protein mediates ESCRT-independent membrane scission
- PMID: 20850012
- PMCID: PMC3059587
- DOI: 10.1016/j.cell.2010.08.029
Influenza virus M2 protein mediates ESCRT-independent membrane scission
Abstract
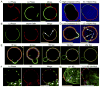
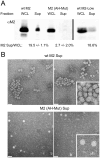
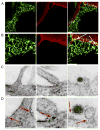
Many viruses utilize host ESCRT proteins for budding; however, influenza virus budding is thought to be ESCRT-independent. In this study we have found a role for the influenza virus M2 proton-selective ion channel protein in mediating virus budding. We observed that a highly conserved amphipathic helix located within the M2 cytoplasmic tail mediates a cholesterol-dependent alteration in membrane curvature. The 17 amino acid amphipathic helix is sufficient for budding into giant unilamellar vesicles, and mutation of this sequence inhibited budding of transfected M2 protein in vivo. We show that M2 localizes to the neck of budding virions and that mutation of the M2 amphipathic helix results in failure of the virus to undergo membrane scission and virion release. These data suggest that M2 mediates the final steps of budding for influenza viruses, bypassing the need for host ESCRT proteins.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Influenza exits the cell without an ESCRT.Cell. 2010 Sep 17;142(6):839-41. doi: 10.1016/j.cell.2010.08.036. Cell. 2010. PMID: 20850005
-
Severing the bud.Nat Rev Microbiol. 2010 Nov;8(11):757. doi: 10.1038/nrmicro2458. Nat Rev Microbiol. 2010. PMID: 21080549 No abstract available.
References
-
- Allain JM, Ben Amar M. Budding and fission of a multiphase vesicle. Eur Phys J E. 2006;20:409–420. - PubMed
-
- Antonny B. Membrane deformation by protein coats. Curr Opin Cell Biol. 2006;18:386–394. - PubMed
-
- Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

