Serpin-1 splicing isoform J inhibits the proSpätzle-activating proteinase HP8 to regulate expression of antimicrobial hemolymph proteins in Manduca sexta
- PMID: 20851714
- PMCID: PMC2956776
- DOI: 10.1016/j.dci.2010.09.004
Serpin-1 splicing isoform J inhibits the proSpätzle-activating proteinase HP8 to regulate expression of antimicrobial hemolymph proteins in Manduca sexta
Abstract
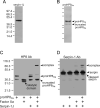
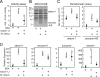
The innate immune system of insects include the Toll pathway, which is mediated by an extracellular serine proteinase cascade. In the tobacco hornworm, Manduca sexta, hemolymph proteinase 8 (HP8) promotes the synthesis of antimicrobial proteins by cleaving proSpätzle, the putative ligand of M. sexta Toll. HP8 has been observed to form a complex in hemolymph with M. sexta serpin-1, which has multiple alternative splicing isoforms. To investigate the regulation of HP8 and its processing of proSpätzle, we characterized the interaction of recombinant HP8 with serpin-1 isoform J (serpin-1J). Recombinant serpin-1J formed an SDS-stable complex with HP8 in vitro. The association rate constant of serpin-1J and HP8 was 1.3×10(4)M(-1)s(-1), with a stoichiometry of inhibition of 5.4. Serpin-1J inhibited the cleavage of proSpätzle by HP8. Injection of serpin-1J into M. sexta larvae resulted in decreased bacteria-induced antimicrobial activity in hemolymph and reduced expression of cecropin, attacin and hemolin mRNA in fat body. Altogether, these results suggest that serpin-1J functions to inhibit HP8 and thereby modulates the concentration of active Spätzle to regulate the Toll pathway response in M. sexta.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Manduca sexta serpin-5 regulates prophenoloxidase activation and the Toll signaling pathway by inhibiting hemolymph proteinase HP6.Insect Biochem Mol Biol. 2010 Sep;40(9):683-9. doi: 10.1016/j.ibmb.2010.07.001. Epub 2010 Jul 17. Insect Biochem Mol Biol. 2010. PMID: 20624461 Free PMC article.
-
Proteolytic activation and function of the cytokine Spätzle in the innate immune response of a lepidopteran insect, Manduca sexta.FEBS J. 2010 Jan;277(1):148-62. doi: 10.1111/j.1742-4658.2009.07465.x. Epub 2009 Nov 26. FEBS J. 2010. PMID: 19968713 Free PMC article.
-
Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways.J Biol Chem. 2009 Jul 17;284(29):19716-26. doi: 10.1074/jbc.M109.007112. Epub 2009 Jun 1. J Biol Chem. 2009. PMID: 19487692 Free PMC article.
-
Innate immune responses of a lepidopteran insect, Manduca sexta.Immunol Rev. 2004 Apr;198:97-105. doi: 10.1111/j.0105-2896.2004.0121.x. Immunol Rev. 2004. PMID: 15199957 Review.
-
Pattern recognition proteins in Manduca sexta plasma.Insect Biochem Mol Biol. 2002 Oct;32(10):1287-93. doi: 10.1016/s0965-1748(02)00091-7. Insect Biochem Mol Biol. 2002. PMID: 12225919 Review.
Cited by
-
Manduca sexta hemolymph protease-1, activated by an unconventional non-proteolytic mechanism, mediates immune responses.Insect Biochem Mol Biol. 2017 May;84:23-31. doi: 10.1016/j.ibmb.2017.03.008. Epub 2017 Mar 31. Insect Biochem Mol Biol. 2017. PMID: 28366787 Free PMC article.
-
Innate Immunity in Insects: The Lights and Shadows of Phenoloxidase System Activation.Int J Mol Sci. 2025 Feb 4;26(3):1320. doi: 10.3390/ijms26031320. Int J Mol Sci. 2025. PMID: 39941087 Free PMC article. Review.
-
Molecular Characterization and Bioinformatics Analysis of a Prophenoloxidase-1 (PPO1) in Plutella xylostella.Int J Insect Sci. 2016 Mar 2;8:1-8. doi: 10.4137/IJIS.S36246. eCollection 2016. Int J Insect Sci. 2016. PMID: 26966394 Free PMC article.
-
Serpin functions in host-pathogen interactions.PeerJ. 2018 Apr 5;6:e4557. doi: 10.7717/peerj.4557. eCollection 2018. PeerJ. 2018. PMID: 29632742 Free PMC article.
-
A Comprehensive Phylogenetic Analysis of the Serpin Superfamily.Mol Biol Evol. 2021 Jun 25;38(7):2915-2929. doi: 10.1093/molbev/msab081. Mol Biol Evol. 2021. PMID: 33744972 Free PMC article.
References
-
- Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–74. - PubMed
-
- Shin SW, Bian G, Raikhel AS. A Toll receptor and a cytokine, Toll5A and Spz1C, are involved in Toll antifungal immune signaling in the mosquito Aedes aegypti. J Biol Chem. 2006;281:39388–95. - PubMed
-
- Kim CH, Kim SJ, Kan H, Kwon HM, Roh KB, Jiang R, et al. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced Toll pathway in an insect. J Biol Chem. 2008;283:7599–607. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

