A novel p53 phosphorylation site within the MDM2 ubiquitination signal: I. phosphorylation at SER269 in vivo is linked to inactivation of p53 function
- PMID: 20851891
- PMCID: PMC2988381
- DOI: 10.1074/jbc.M110.143099
A novel p53 phosphorylation site within the MDM2 ubiquitination signal: I. phosphorylation at SER269 in vivo is linked to inactivation of p53 function
Abstract
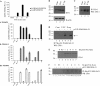
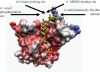
p53 is a thermodynamically unstable protein containing a conformationally flexible multiprotein docking site within the DNA-binding domain. A combinatorial peptide chip used to identify the novel kinase consensus site RXSΦ(K/D) led to the discovery of a homologous phosphorylation site in the S10 β-strand of p53 at Ser(269). Overlapping peptide libraries confirmed that Ser(269) was a phosphoacceptor site in vitro, and immunochemical approaches evaluated whether p53 is phosphorylated in vivo at Ser(269). Mutation or phosphorylation of p53 at Ser(269) attenuates binding of the p53-specific monoclonal antibody DO-12, identifying an assay for measuring Ser(269) phosphorylation of p53 in vivo. The mAb DO-12 epitope of p53 is masked via phosphorylation in a range of human tumor cells with WT p53 status, as defined by increased mAb DO-12 binding to endogenous p53 after phosphatase treatment. Phospho-Ser(269)-specific monoclonal antibodies were generated and used to demonstrate that p53 phosphorylation is induced at Ser(269) after irradiation with kinetics similar to those of p53 protein induction. Phosphomimetic mutation at Ser(269) inactivated the transcription activation function and clonogenic suppressor activity of p53. These data suggest that the dynamic equilibrium between native and unfolded states of WT p53 can be modulated by phosphorylation of the conformationally flexible multiprotein binding site in the p53 DNA-binding domain.
Figures






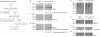


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

