Predicting plasmid promiscuity based on genomic signature
- PMID: 20851899
- PMCID: PMC2976448
- DOI: 10.1128/JB.00277-10
Predicting plasmid promiscuity based on genomic signature
Abstract
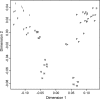
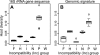
Despite the important contribution of self-transmissible plasmids to bacterial evolution, little is understood about the range of hosts in which these plasmids have evolved. Our goal was to infer this so-called evolutionary host range. The nucleotide composition, or genomic signature, of plasmids is often similar to that of the chromosome of their current host, suggesting that plasmids acquire their hosts' signature over time. Therefore, we examined whether the evolutionary host range of plasmids could be inferred by comparing their trinucleotide composition to that of all completely sequenced bacterial chromosomes. The diversity of candidate hosts was determined using taxonomic classification and genetic distance. The method was first tested using plasmids from six incompatibility (Inc) groups whose host ranges are generally thought to be narrow (IncF, IncH, and IncI) or broad (IncN, IncP, and IncW) and then applied to other plasmid groups. The evolutionary host range was found to be broad for IncP plasmids, narrow for IncF and IncI plasmids, and intermediate for IncH and IncN plasmids, which corresponds with their known host range. The IncW plasmids as well as several plasmids from the IncA/C, IncP, IncQ, IncU, and PromA groups have signatures that were not similar to any of the chromosomal signatures, raising the hypothesis that these plasmids have not been ameliorated in any host due to their promiscuous nature. The inferred evolutionary host range of IncA/C, IncP-9, and IncL/M plasmids requires further investigation. In this era of high-throughput sequencing, this genomic signature method is a useful tool for predicting the host range of novel mobile elements.
Figures


Similar articles
-
Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR.Appl Environ Microbiol. 1996 Jul;62(7):2621-8. doi: 10.1128/aem.62.7.2621-2628.1996. Appl Environ Microbiol. 1996. PMID: 8779598 Free PMC article.
-
IncP-1 and PromA group plasmids are major providers of horizontal gene transfer capacities across bacteria in the mycosphere of different soil fungi.Microb Ecol. 2015 Jan;69(1):169-79. doi: 10.1007/s00248-014-0482-6. Epub 2014 Aug 23. Microb Ecol. 2015. PMID: 25149284
-
A survey of five broad-host-range plasmids in gram-negative bacilli isolated from patients.Plasmid. 2014 Jul;74:9-14. doi: 10.1016/j.plasmid.2014.05.002. Epub 2014 May 24. Plasmid. 2014. PMID: 24864033
-
Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae.J Antimicrob Chemother. 2018 May 1;73(5):1121-1137. doi: 10.1093/jac/dkx488. J Antimicrob Chemother. 2018. PMID: 29370371 Review.
-
Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution.FEMS Microbiol Rev. 2006 Nov;30(6):942-66. doi: 10.1111/j.1574-6976.2006.00042.x. Epub 2006 Oct 6. FEMS Microbiol Rev. 2006. PMID: 17026718 Review.
Cited by
-
Plasmid Detection, Characterization, and Ecology.Microbiol Spectr. 2015 Feb;3(1):PLAS-0038-2014. doi: 10.1128/microbiolspec.PLAS-0038-2014. Microbiol Spectr. 2015. PMID: 26104560 Free PMC article. Review.
-
Construction of a shuttle expression vector for lactic acid bacteria.J Genet Eng Biotechnol. 2019 Nov 18;17(1):10. doi: 10.1186/s43141-019-0013-4. J Genet Eng Biotechnol. 2019. PMID: 31736018 Free PMC article.
-
Associations between multidrug resistance, plasmid content, and virulence potential among extraintestinal pathogenic and commensal Escherichia coli from humans and poultry.Foodborne Pathog Dis. 2012 Jan;9(1):37-46. doi: 10.1089/fpd.2011.0961. Epub 2011 Oct 11. Foodborne Pathog Dis. 2012. PMID: 21988401 Free PMC article.
-
Conjugative transfer of an IncA/C plasmid-borne blaCMY-2 gene through genetic re-arrangements with an IncX1 plasmid.BMC Microbiol. 2013 Nov 21;13:264. doi: 10.1186/1471-2180-13-264. BMC Microbiol. 2013. PMID: 24262067 Free PMC article.
-
Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance.Antimicrob Agents Chemother. 2011 Aug;55(8):3649-60. doi: 10.1128/AAC.00013-11. Epub 2011 May 16. Antimicrob Agents Chemother. 2011. PMID: 21576439 Free PMC article. Review.
References
-
- Arakawa, K., K. Mori, K. Ikeda, T. Matsuzaki, Y. Kobayashi, and M. Tomita. 2003. G-language genome analysis environment: a workbench for nucleotide sequence data mining. Bioinformatics 19:305-306. - PubMed
-
- Asano, K., and K. Mizobuchi. 1998. An RNA pseudoknot as the molecular switch for translation of the repZ gene encoding the replication initiator of IncIalpha plasmid ColIb-P9. J. Biol. Chem. 273:11815-11825. - PubMed
-
- Bahl, M. I., L. H. Hansen, A. Goesmann, and S. J. Sorensen. 2007. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established alpha, beta and delta sub-groups. Plasmid 58:31-43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

