Cell wall anchoring of the 37-kilodalton oncofetal antigen by Lactobacillus plantarum for mucosal cancer vaccine delivery
- PMID: 20851975
- PMCID: PMC2976233
- DOI: 10.1128/AEM.01031-10
Cell wall anchoring of the 37-kilodalton oncofetal antigen by Lactobacillus plantarum for mucosal cancer vaccine delivery
Abstract
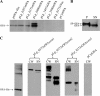
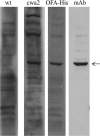
The 37-kDa oncofetal antigen (OFA), a tumor immunogen expressed on all mammalian cancers examined to date, was secreted and anchored to the cell wall of Lactobacillus plantarum using homologous signal peptides and LPxTG anchors. Orally administered L. plantarum expressing anchored OFA induced a specific immune response against OFA in mice.
Figures

Similar articles
-
Recombinant Lactobacillus plantarum induces immune responses to cancer testis antigen NY-ESO-1 and maturation of dendritic cells.Hum Vaccin Immunother. 2015;11(11):2664-73. doi: 10.1080/21645515.2015.1056952. Epub 2015 Jul 17. Hum Vaccin Immunother. 2015. PMID: 26185907 Free PMC article.
-
Anchoring of heterologous proteins in multiple Lactobacillus species using anchors derived from Lactobacillus plantarum.Sci Rep. 2020 Jun 15;10(1):9640. doi: 10.1038/s41598-020-66531-7. Sci Rep. 2020. PMID: 32541679 Free PMC article.
-
Production, safety and antitumor efficacy of recombinant Oncofetal Antigen/immature laminin receptor protein.Biomaterials. 2009 Jun;30(17):3091-9. doi: 10.1016/j.biomaterials.2009.02.022. Epub 2009 Mar 6. Biomaterials. 2009. PMID: 19268360
-
Immunogenic Properties of Lactobacillus plantarum Producing Surface-Displayed Mycobacterium tuberculosis Antigens.Appl Environ Microbiol. 2016 Dec 30;83(2):e02782-16. doi: 10.1128/AEM.02782-16. Print 2017 Jan 15. Appl Environ Microbiol. 2016. PMID: 27815271 Free PMC article.
-
The benefits of Lactiplantibacillus plantarum: From immunomodulator to vaccine vector.Immunol Lett. 2025 Apr;272:106971. doi: 10.1016/j.imlet.2025.106971. Epub 2025 Jan 5. Immunol Lett. 2025. PMID: 39765312 Review.
Cited by
-
Mucosal and systemic immune responses induced by recombinant Lactobacillus spp. expressing the hemagglutinin of the avian influenza virus H5N1.Clin Vaccine Immunol. 2012 Feb;19(2):174-9. doi: 10.1128/CVI.05618-11. Epub 2011 Nov 30. Clin Vaccine Immunol. 2012. PMID: 22131355 Free PMC article.
-
Antigen surface display in two novel whole genome sequenced food grade strains, Lactiplantibacillus pentosus KW1 and KW2.Microb Cell Fact. 2024 Jan 11;23(1):19. doi: 10.1186/s12934-024-02296-2. Microb Cell Fact. 2024. PMID: 38212746 Free PMC article.
-
Effective treatment of hypertension by recombinant Lactobacillus plantarum expressing angiotensin converting enzyme inhibitory peptide.Microb Cell Fact. 2015 Dec 21;14:202. doi: 10.1186/s12934-015-0394-2. Microb Cell Fact. 2015. PMID: 26691527 Free PMC article. Clinical Trial.
-
Surface display of glycosylated Tyrosinase related protein-2 (TRP-2) tumour antigen on Lactococcus lactis.BMC Biotechnol. 2015 Dec 29;15:113. doi: 10.1186/s12896-015-0231-z. BMC Biotechnol. 2015. PMID: 26715153 Free PMC article.
-
A SH3_5 Cell Anchoring Domain for Non-recombinant Surface Display on Lactic Acid Bacteria.Front Bioeng Biotechnol. 2021 Jan 27;8:614498. doi: 10.3389/fbioe.2020.614498. eCollection 2020. Front Bioeng Biotechnol. 2021. PMID: 33585415 Free PMC article.
References
-
- Aires, K. A., A. M. Cianciarullo, S. M. Carneiro, L. L. Villa, E. Boccardo, G. Perez-Martinez, I. Perez-Arellano, M. L. Oliveira, and P. L. Ho. 2006. Production of human papillomavirus type 16 L1 virus-like particles by recombinant Lactobacillus casei cells. Appl. Environ. Microbiol. 72:745-752. - PMC - PubMed
-
- Aslakson, C. J., and F. R. Miller. 1992. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 52:1399-1405. - PubMed
-
- Aukrust, T. W., M. B. Brurberg, and I. F. Nes. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201-208. - PubMed
-
- Barsoum, A. L., J. W. Rohrer, and J. H. Coggin. 2000. 37kDa oncofetal antigen is an autoimmunogenic homologue of the 37kDa laminin receptor precursor. Cell. Mol. Biol. Lett. 5:207-230.
-
- Bermudez-Humaran, L. G., N. G. Cortes-Perez, F. Lefevre, V. Guimaraes, S. Rabot, J. M. Alcocer-Gonzalez, J. J. Gratadoux, C. Rodriguez-Padilla, R. S. Tamez-Guerra, G. Corthier, A. Gruss, and P. Langella. 2005. A novel mucosal vaccine based on live lactococci expressing E7 antigen and IL-12 induces systemic and mucosal immune responses and protects mice against human papillomavirus type 16-induced tumors. J. Immunol. 175:7297-7302. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

