The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex
- PMID: 20852019
- PMCID: PMC2965543
- DOI: 10.1105/tpc.110.076406
The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex
Abstract
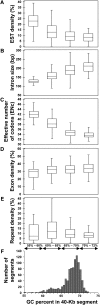
Chlorella variabilis NC64A, a unicellular photosynthetic green alga (Trebouxiophyceae), is an intracellular photobiont of Paramecium bursaria and a model system for studying virus/algal interactions. We sequenced its 46-Mb nuclear genome, revealing an expansion of protein families that could have participated in adaptation to symbiosis. NC64A exhibits variations in GC content across its genome that correlate with global expression level, average intron size, and codon usage bias. Although Chlorella species have been assumed to be asexual and nonmotile, the NC64A genome encodes all the known meiosis-specific proteins and a subset of proteins found in flagella. We hypothesize that Chlorella might have retained a flagella-derived structure that could be involved in sexual reproduction. Furthermore, a survey of phytohormone pathways in chlorophyte algae identified algal orthologs of Arabidopsis thaliana genes involved in hormone biosynthesis and signaling, suggesting that these functions were established prior to the evolution of land plants. We show that the ability of Chlorella to produce chitinous cell walls likely resulted from the capture of metabolic genes by horizontal gene transfer from algal viruses, prokaryotes, or fungi. Analysis of the NC64A genome substantially advances our understanding of the green lineage evolution, including the genomic interplay with viruses and symbiosis between eukaryotes.
Figures




Comment in
-
The Chlorella genome: big surprises from a small package.Plant Cell. 2010 Sep;22(9):2924. doi: 10.1105/tpc.110.220911. Epub 2010 Sep 17. Plant Cell. 2010. PMID: 20852020 Free PMC article. No abstract available.
References
-
- Aparicio S., et al. (2002). Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310 - PubMed
-
- Bajguz A. (2009). Brassinosteroid enhanced the level of abscisic acid in Chlorella vulgaris subjected to short-term heat stress. J. Plant Physiol. 166: 882–886 - PubMed
-
- Bateman R.M., Crane P.R., DiMichele W.A., Kenrick P.R., Rowe N.P., Speck T., Stein W.E. (1998). Early evolution of land plants: phylogeny, physiology, and ecology of the primary terrestrial radiation. Annu. Rev. Ecol. Syst. 29: 263–292
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

