Aspergillus fumigatus flbB encodes two basic leucine zipper domain (bZIP) proteins required for proper asexual development and gliotoxin production
- PMID: 20852021
- PMCID: PMC2976297
- DOI: 10.1128/EC.00198-10
Aspergillus fumigatus flbB encodes two basic leucine zipper domain (bZIP) proteins required for proper asexual development and gliotoxin production
Abstract
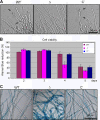
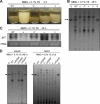
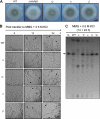
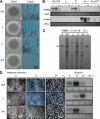
The opportunistic human pathogen Aspergillus fumigatus reproduces asexually by forming a massive number of mitospores called conidia. In this study, we characterize the upstream developmental regulator A. fumigatus flbB (AfuflbB). Northern blotting and cDNA analyses reveal that AfuflbB produces two transcripts predicted to encode two basic leucine zipper domain (bZIP) polypeptides, AfuFlbBβ (420 amino acids [aa]) and AfuFlbBα (390 aa). The deletion of AfuflbB results in delayed/reduced sporulation, precocious cell death, the lack of conidiophore development in liquid submerged culture, altered expression of AfubrlA and AfuabaA, and blocked production of gliotoxin. While introduction of the wild-type (WT) AfuflbB allele fully complemented these defects, disruption of the ATG start codon for either one of the AfuFlbB polypeptides leads to a partial complementation, indicating the need of both polypeptides for WT levels of asexual development and gliotoxin biogenesis. Consistent with this, Aspergillus nidulans flbB(+) encoding one polypeptide (426 aa) partially complements the AfuflbB null mutation. The presence of 0.6 M KCl in liquid submerged culture suppresses the defects caused by the lack of one, but not both, of the AfuFlbB polypeptides, suggesting a genetic prerequisite for AfuFlbB in A. fumigatus development. Finally, Northern blot analyses reveal that both AfuflbB and AfuflbE are necessary for expression of AfuflbD, suggesting that FlbD functions downstream of FlbB/FlbE in aspergilli.
Figures




Similar articles
-
Characterization of the developmental regulator FlbE in Aspergillus fumigatus and Aspergillus nidulans.Fungal Genet Biol. 2010 Dec;47(12):981-93. doi: 10.1016/j.fgb.2010.08.009. Epub 2010 Sep 9. Fungal Genet Biol. 2010. PMID: 20817115
-
The developmental regulators, FlbB and FlbE, are involved in the virulence of Aspergillus fumigatus.J Microbiol Biotechnol. 2013 Jun 28;23(6):766-70. doi: 10.4014/jmb.1301.01002. J Microbiol Biotechnol. 2013. PMID: 23676908
-
Basic-zipper-type transcription factor FlbB controls asexual development in Aspergillus nidulans.Eukaryot Cell. 2008 Jan;7(1):38-48. doi: 10.1128/EC.00207-07. Epub 2007 Nov 9. Eukaryot Cell. 2008. PMID: 17993569 Free PMC article.
-
Genetic control of asexual development in aspergillus fumigatus.Adv Appl Microbiol. 2015;90:93-107. doi: 10.1016/bs.aambs.2014.09.004. Epub 2014 Nov 12. Adv Appl Microbiol. 2015. PMID: 25596030 Review.
-
Developmental regulators in Aspergillus fumigatus.J Microbiol. 2016 Mar;54(3):223-31. doi: 10.1007/s12275-016-5619-5. Epub 2016 Feb 27. J Microbiol. 2016. PMID: 26920882 Review.
Cited by
-
Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum.Genes (Basel). 2022 Mar 28;13(4):607. doi: 10.3390/genes13040607. Genes (Basel). 2022. PMID: 35456413 Free PMC article.
-
Research on the Molecular Interaction Mechanism between Plants and Pathogenic Fungi.Int J Mol Sci. 2022 Apr 22;23(9):4658. doi: 10.3390/ijms23094658. Int J Mol Sci. 2022. PMID: 35563048 Free PMC article. Review.
-
Regulation of Conidiogenesis in Aspergillus flavus.Cells. 2022 Sep 7;11(18):2796. doi: 10.3390/cells11182796. Cells. 2022. PMID: 36139369 Free PMC article. Review.
-
Systematic Characterization of bZIP Transcription Factors Required for Development and Aflatoxin Generation by High-Throughput Gene Knockout in Aspergillus flavus.J Fungi (Basel). 2022 Mar 30;8(4):356. doi: 10.3390/jof8040356. J Fungi (Basel). 2022. PMID: 35448587 Free PMC article.
-
Development in Aspergillus.Stud Mycol. 2013 Mar 15;74(1):1-29. doi: 10.3114/sim0006. Epub 2012 Sep 14. Stud Mycol. 2013. PMID: 23450714 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

