Characterization of glycoside hydrolase family 5 proteins in Schizosaccharomyces pombe
- PMID: 20852022
- PMCID: PMC2976300
- DOI: 10.1128/EC.00187-10
Characterization of glycoside hydrolase family 5 proteins in Schizosaccharomyces pombe
Abstract
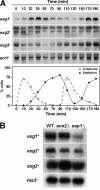
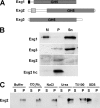
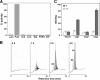
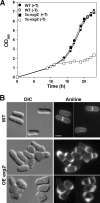
In yeast, enzymes with β-glucanase activity are thought to be necessary in morphogenetic events that require controlled hydrolysis of the cell wall. Comparison of the sequence of the Saccharomyces cerevisiae exo-β(1,3)-glucanase Exg1 with the Schizosaccharomyces pombe genome allowed the identification of three genes that were named exg1(+) (locus SPBC1105.05), exg2(+) (SPAC12B10.11), and exg3(+) (SPBC2D10.05). The three proteins have different localizations: Exg1 is secreted to the periplasmic space, Exg2 is a membrane protein, and Exg3 is a cytoplasmic protein. Characterization of the biochemical activity of the proteins indicated that Exg1 and Exg3 are active only against β(1,6)-glucans while no activity was detected for Exg2. Interestingly, Exg1 cleaves the glucans with an endohydrolytic mode of action. exg1(+) showed periodic expression during the cell cycle, with a maximum coinciding with the septation process, and its expression was dependent on the transcription factor Sep1. The Exg1 protein localizes to the septum region in a pattern that was different from that of the endo-β(1,3)-glucanase Eng1. Overexpression of Exg2 resulted in an increase in cell wall material at the poles and in the septum, but the putative catalytic activity of the protein was not required for this effect.
Figures


Similar articles
-
The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe.J Cell Sci. 2003 May 1;116(Pt 9):1689-98. doi: 10.1242/jcs.00377. J Cell Sci. 2003. PMID: 12665550
-
The Schizosaccharomyces pombe endo-1,3-beta-glucanase Eng1 contains a novel carbohydrate binding module required for septum localization.Mol Microbiol. 2008 Jul;69(1):188-200. doi: 10.1111/j.1365-2958.2008.06275.x. Epub 2008 May 5. Mol Microbiol. 2008. PMID: 18466295
-
Role of the alpha-glucanase Agn1p in fission-yeast cell separation.Mol Biol Cell. 2004 Aug;15(8):3903-14. doi: 10.1091/mbc.e04-04-0319. Epub 2004 Jun 11. Mol Biol Cell. 2004. PMID: 15194814 Free PMC article.
-
Splitting of the fission yeast septum.FEMS Yeast Res. 2007 Sep;7(6):761-70. doi: 10.1111/j.1567-1364.2007.00266.x. Epub 2007 Jun 27. FEMS Yeast Res. 2007. PMID: 17596184 Review.
-
Using genetics to understand biology.Heredity (Edinb). 2019 Jul;123(1):4-13. doi: 10.1038/s41437-019-0209-z. Epub 2019 Jun 12. Heredity (Edinb). 2019. PMID: 31189902 Free PMC article. Review. No abstract available.
Cited by
-
Ultrastructural plasma membrane asymmetries in tension and curvature promote yeast cell fusion.J Cell Biol. 2021 Oct 4;220(10):e202103142. doi: 10.1083/jcb.202103142. Epub 2021 Aug 12. J Cell Biol. 2021. PMID: 34382996 Free PMC article.
-
Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5).BMC Evol Biol. 2012 Sep 20;12:186. doi: 10.1186/1471-2148-12-186. BMC Evol Biol. 2012. PMID: 22992189 Free PMC article.
-
A formin-nucleated actin aster concentrates cell wall hydrolases for cell fusion in fission yeast.J Cell Biol. 2015 Mar 30;208(7):897-911. doi: 10.1083/jcb.201411124. J Cell Biol. 2015. PMID: 25825517 Free PMC article.
-
β-1,3-glucan modifying enzymes in Aspergillus fumigatus.Front Microbiol. 2013 Apr 17;4:81. doi: 10.3389/fmicb.2013.00081. eCollection 2013. Front Microbiol. 2013. PMID: 23616783 Free PMC article.
-
Off the wall: The rhyme and reason of Neurospora crassa hyphal morphogenesis.Cell Surf. 2019 Mar 8;5:100020. doi: 10.1016/j.tcsw.2019.100020. eCollection 2019 Dec. Cell Surf. 2019. PMID: 32743136 Free PMC article. Review.
References
-
- Adams D. J. 2004. Fungal cell wall chitinases and glucanases. Microbiology 150:2029–2035 - PubMed
-
- Arellano M., Duran A., Perez P. 1997. Localisation of the Schizosaccharomyces pombe rho1p GTPase and its involvement in the organisation of the actin cytoskeleton. J. Cell Sci. 110:2547–2555 - PubMed
-
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., Steever A. B., Wach A., Philippsen P., Pringle J. R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

