Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1
- PMID: 20852131
- PMCID: PMC3031398
- DOI: 10.1182/blood-2010-05-286328
Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1
Abstract
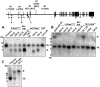
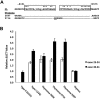
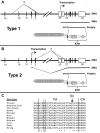
Point mutations that trigger ligand-independent proteolysis of the Notch1 ectodomain occur frequently in human T-cell acute lymphoblastic leukemia (T-ALL) but are rare in murine T-ALL, suggesting that other mechanisms account for Notch1 activation in murine tumors. Here we show that most murine T-ALLs harbor Notch1 deletions that fall into 2 types, both leading to ligand-independent Notch1 activation. Type 1 deletions remove exon 1 and the proximal promoter, appear to be RAG-mediated, and are associated with mRNA transcripts that initiate from 3' regions of Notch1. In line with the RAG dependency of these rearrangements, RAG2 binds to the 5' end of Notch1 in normal thymocytes near the deletion breakpoints. Type 2 deletions remove sequences between exon 1 and exons 26 to 28 of Notch1, appear to be RAG-independent, and are associated with transcripts in which exon 1 is spliced out of frame to 3' Notch1 exons. Translation of both types of transcripts initiates at a conserved methionine residue, M1727, which lies within the Notch1 transmembrane domain. Polypeptides initiating at M1727 insert into membranes and are subject to constitutive cleavage by γ-secretase. Thus, like human T-ALL, murine T-ALL is often associated with acquired mutations that cause ligand-independent Notch1 activation.
Figures




Comment in
-
"Cryptic" Notch1 messages induce T-ALL.Blood. 2010 Dec 16;116(25):5436-8. doi: 10.1182/blood-2010-10-311712. Blood. 2010. PMID: 21163932 No abstract available.
References
-
- Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. - PubMed
-
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14(4):295–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

