Phenotypic effects of Ehlers-Danlos syndrome-associated mutation on the FnIII domain of tenascin-X
- PMID: 20853426
- PMCID: PMC3005793
- DOI: 10.1002/pro.503
Phenotypic effects of Ehlers-Danlos syndrome-associated mutation on the FnIII domain of tenascin-X
Abstract
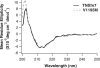
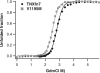
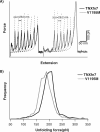
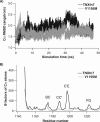
Tenascin-X (TNX) is an extracellular matrix (ECM) protein and interacts with a wide variety of molecules in the ECM as well as on the membrane. Deficiency of TNX causes a recessive form of Ehlers-Danlos syndrome (EDS) characterized by hyperelastic and fragile skin, easy bruising, and hypermobile joints. Three point mutations in TNX gene were found to be associated with hypermobility type EDS and one of such mutations is the V1195M mutation at the 7th fibronectin Type III domain (TNXfn7). To help elucidate the underlying molecular mechanism connecting this mutation to EDS, here we combined homology modeling, chemical denaturation, single molecule atomic force microscopy, and molecular dynamics (MD) simulation techniques to investigate the phenotypic effects of V1195M on TNXfn7. We found that the V1195M mutation does not alter the three-dimensional structure of TNXfn7 and had only mild destabilization effects on the thermodynamic and mechanical stability of TNXfn7. However, MD simulations revealed that the mutation V1195M significantly alters the flexibility of the C'E loop of TNXfn7. As loops play important roles in protein-protein and protein-ligand interactions, we hypothesize that the decreased loop flexibility by V1195M mutation may affect the binding of TNX to ECM molecules and thus adversely affect collagen deposition and fibrillogenesis. Our results may provide new insights in understanding the molecular basis for the pathogenesis of V1195M-resulted EDS.
Figures

Similar articles
-
Elastic fiber abnormalities in hypermobility type Ehlers-Danlos syndrome patients with tenascin-X mutations.Clin Genet. 2005 Apr;67(4):330-4. doi: 10.1111/j.1399-0004.2005.00401.x. Clin Genet. 2005. PMID: 15733269
-
A recessive form of the Ehlers-Danlos syndrome caused by tenascin-X deficiency.N Engl J Med. 2001 Oct 18;345(16):1167-75. doi: 10.1056/NEJMoa002939. N Engl J Med. 2001. PMID: 11642233
-
Clinical and Molecular Characterization of Classical-Like Ehlers-Danlos Syndrome Due to a Novel TNXB Variant.Genes (Basel). 2019 Oct 25;10(11):843. doi: 10.3390/genes10110843. Genes (Basel). 2019. PMID: 31731524 Free PMC article.
-
Tenascin-X, Congenital Adrenal Hyperplasia, and the CAH-X Syndrome.Horm Res Paediatr. 2018;89(5):352-361. doi: 10.1159/000481911. Epub 2018 May 7. Horm Res Paediatr. 2018. PMID: 29734195 Free PMC article. Review.
-
Tenascin-X, collagen, elastin, and the Ehlers-Danlos syndrome.Am J Med Genet C Semin Med Genet. 2005 Nov 15;139C(1):24-30. doi: 10.1002/ajmg.c.30071. Am J Med Genet C Semin Med Genet. 2005. PMID: 16278880 Review.
Cited by
-
Disease variants in genomes of 44 centenarians.Mol Genet Genomic Med. 2014 Sep;2(5):438-50. doi: 10.1002/mgg3.86. Epub 2014 Jun 15. Mol Genet Genomic Med. 2014. PMID: 25333069 Free PMC article.
References
-
- Chiquet-Ehrismann R. Tenascins, a growing family of extracellular matrix proteins. Experientia. 1995;51:853–862. - PubMed
-
- Elefteriou F, Exposito JY, Garrone R, Lethias C. Characterization of the bovine tenascin-X. J Biol Chem. 1997;272:22866–22874. - PubMed
-
- Erickson HP. Tenascin-C, tenascin-R and tenascin-X: a family of talented proteins in search of functions. Curr Opin Cell Biol. 1993;5:869–876. - PubMed
-
- Jones FS, Jones PL. The tenascin family of ECM glycoproteins: structure, function, and regulation during embryonic development and tissue remodeling. Dev Dyn. 2000;218:235–259. - PubMed
-
- Elefteriou F, Exposito JY, Garrone R, Lethias C. Binding of tenascin-X to decorin. FEBS Lett. 2001;495:44–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

