Molecular characterization of tlyA gene product, Rv1694 of Mycobacterium tuberculosis: a non-conventional hemolysin and a ribosomal RNA methyl transferase
- PMID: 20854656
- PMCID: PMC2954847
- DOI: 10.1186/1471-2091-11-35
Molecular characterization of tlyA gene product, Rv1694 of Mycobacterium tuberculosis: a non-conventional hemolysin and a ribosomal RNA methyl transferase
Abstract
Background: Mycobacterium tuberculosis is a virulent bacillus causing tuberculosis, a disease responsible for million deaths each year worldwide. In order to understand its mechanism of pathogenesis in humans and to help control tuberculosis, functions of numerous Mycobacterium tuberculosis genes are being characterized. In this study we report the dual functionality of tlyA gene product of Mycobacterium tuberculosis annotated as Rv1694, a 268 amino acid long basic protein.
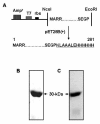
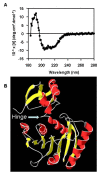
Results: The recombinant purified Rv1694 protein was found to exhibit hemolytic activity in vitro. It showed concentration and time-dependent hemolysis of rabbit and human erythrocytes. Multiple oligomeric forms (dimers to heptamers) of this protein were seen on the membranes of the lysed erythrocytes. Like the oligomers of conventional, well-known, pore-forming toxins, the oligomers of Rv1694 were found to be resistant to heat and SDS, but were susceptible to reducing agents like β-mercaptoethanol as it had abolished the hemolytic activity of Rv1694 indicating the role of disulfide bond(s). The Rv1694 generated de novo by in vitro transcription and translation also exhibited unambiguous hemolysis confirming the self assembly and oligomerization properties of this protein. Limited proteolytic digestion of this protein has revealed that the amino terminus is susceptible while in solution but is protected in presence of membrane. Striking feature of Rv1694 is its presence on the cell wall of E. coli as visualized by confocal microscopy. The surface expression is consistent with the contact dependent haemolytic ability of E. coli expressing this protein. Also, immune serum specific to this protein inhibits the contact dependent hemolysis. Moreover, Rv1694 protein binds to and forms stable oligomers on the macrophage phagosomal membranes. In addition to all these properties, E. coli expressing Rv1694 was found to be susceptible to the antibiotic capreomycin as its growth was significantly slower than mock vector transformed E. coli. The S30 extract of E. coli expressing the Rv1694 had poor translational activity in presence of capreomycin, further confirming its methylation activity. Finally, incorporation of methyl group of [3H]-S-adenosylmethionine in isolated ribosomes also confirmed its methylation activity.
Conclusions: The Rv1694 has an unusual dual activity. It appears to contain two diverse functions such as haemolytic activity and ribosomal RNA methylation activity. It is possible that the haemolytic activity might be relevant to intra-cellular compartments such as phagosomes rather than cell lysis of erythrocytes and the self-assembly trait may have a potential role after successful entry into macrophages by Mycobacterium tuberculosis.
Figures






References
-
- Camus JC, Pryor MJ, Medigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. - PubMed
-
- Betts JC, Dodson P, Quan S, Lewis AP, Thomas PJ, Duncan K, McAdam RA. Comparison of the proteome of Mycobacterium tuberculosis strain H37Rv with clinical isolate CDC 1551. Microbiology. 2000;146(Pt 12):3205–3216. - PubMed
-
- Mollenkopf HJ, Jungblut PR, Raupach B, Mattow J, Lamer S, Zimny-Arndt U, Schaible UE, Kaufmann SH. A dynamic two-dimensional polyacrylamide gel electrophoresis database: the mycobacterial proteome via Internet. Electrophoresis. 1999;20:2172–2180. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2172::AID-ELPS2172>3.0.CO;2-M. - DOI - PubMed
-
- Jungblut PR, Schaible UE, Mollenkopf HJ, Zimny-Arndt U, Raupach B, Mattow J, Halada P, Lamer S, Hagens K, Kaufmann SH. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol Microbiol. 1999;33:1103–1117. doi: 10.1046/j.1365-2958.1999.01549.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

