Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill
- PMID: 20854682
- PMCID: PMC2955024
- DOI: 10.1186/1471-2199-11-73
Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill
Abstract
Background: Eucalyptus globulus and its hybrids are very important for the cellulose and paper industry mainly due to their low lignin content and frost resistance. However, rooting of cuttings of this species is recalcitrant and exogenous auxin application is often necessary for good root development. To date one of the most accurate methods available for gene expression analysis is quantitative reverse transcription-polymerase chain reaction (qPCR); however, reliable use of this technique requires reference genes for normalization. There is no single reference gene that can be regarded as universal for all experiments and biological materials. Thus, the identification of reliable reference genes must be done for every species and experimental approach. The present study aimed at identifying suitable control genes for normalization of gene expression associated with adventitious rooting in E. globulus microcuttings.
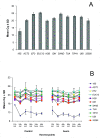
Results: By the use of two distinct algorithms, geNorm and NormFinder, we have assessed gene expression stability of eleven candidate reference genes in E. globulus: 18S, ACT2, EF2, EUC12, H2B, IDH, SAND, TIP41, TUA, UBI and 33380. The candidate reference genes were evaluated in microccuttings rooted in vitro, in presence or absence of auxin, along six time-points spanning the process of adventitious rooting. Overall, the stability profiles of these genes determined with each one of the algorithms were very similar. Slight differences were observed in the most stable pair of genes indicated by each program: IDH and SAND for geNorm, and H2B and TUA for NormFinder. Both programs identified UBI and 18S as the most variable genes. To validate these results and select the most suitable reference genes, the expression profile of the ARGONAUTE1 gene was evaluated in relation to the most stable candidate genes indicated by each algorithm.
Conclusion: Our study showed that expression stability varied between putative reference genes tested in E. globulus. Based on the AGO1 relative expression profile obtained using the genes suggested by the algorithms, H2B and TUA were considered as the most suitable reference genes for expression studies in E. globulus adventitious rooting. UBI and 18S were unsuitable for use as controls in qPCR related to this process. These findings will enable more accurate and reliable normalization of qPCR results for gene expression studies in this economically important woody plant, particularly related to rooting and clonal propagation.
Figures




References
-
- Del Lungo A, Ball J, Carle J. Planted Forests and Trees Working Paper 38. FAO (Food and Agriculture Organization of the United Nations - Forestry Department); 2006. Global planted forests thematic study: results and analysis.
-
- Statistical yearbook from ABRAF. Brazilian Association of Foresters (Associação Brasileira de Produtores de Florestas Plantadas); 2009.
-
- Mora AL, Garcia CH. The Eucalyptus Culture in Brazil (A Cultura do Eucalipto no Brasil) SBS, São Paulo; 2000. p. 112.
-
- Le Roux JJ, Van Staden J. Micropropagation and tissue culture of Eucalyptus - a review. Tree Physiol. 1991;9:435–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

