Structure and mechanism of enzymes involved in biosynthesis and breakdown of the phosphonates fosfomycin, dehydrophos, and phosphinothricin
- PMID: 20854789
- PMCID: PMC3040005
- DOI: 10.1016/j.abb.2010.09.012
Structure and mechanism of enzymes involved in biosynthesis and breakdown of the phosphonates fosfomycin, dehydrophos, and phosphinothricin
Abstract
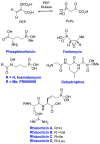
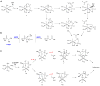
Recent years have seen a rapid increase in the mechanistic and structural information on enzymes that are involved in the biosynthesis and breakdown of naturally occurring phosphonates. This review focuses on these recent developments with an emphasis on those enzymes that have been characterized crystallographically in the past five years, including proteins involved in the biosynthesis of phosphinothricin, fosfomycin, and dehydrophos and proteins involved in resistance mechanisms.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology.Curr Opin Chem Biol. 2013 Aug;17(4):580-8. doi: 10.1016/j.cbpa.2013.06.018. Epub 2013 Jul 17. Curr Opin Chem Biol. 2013. PMID: 23870698 Free PMC article. Review.
-
Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways.J Biol Chem. 2008 Aug 22;283(34):23161-8. doi: 10.1074/jbc.M801788200. Epub 2008 Jun 10. J Biol Chem. 2008. PMID: 18544530 Free PMC article.
-
Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.Proc Natl Acad Sci U S A. 2010 Oct 12;107(41):17557-62. doi: 10.1073/pnas.1006848107. Epub 2010 Sep 27. Proc Natl Acad Sci U S A. 2010. PMID: 20876132 Free PMC article.
-
Fosfomycin induced structural change in fosfomycin resistance kinases FomA: molecular dynamics and molecular docking studies.J Mol Model. 2014 May;20(5):2236. doi: 10.1007/s00894-014-2236-2. Epub 2014 Apr 27. J Mol Model. 2014. PMID: 24770549
-
The Abc of Phosphonate Breakdown: A Mechanism for Bacterial Survival.Bioessays. 2018 Nov;40(11):e1800091. doi: 10.1002/bies.201800091. Epub 2018 Sep 9. Bioessays. 2018. PMID: 30198068 Review.
Cited by
-
The Streptomyces-produced antibiotic fosfomycin is a promiscuous substrate for archaeal isopentenyl phosphate kinase.Biochemistry. 2012 Jan 31;51(4):917-25. doi: 10.1021/bi201662k. Epub 2012 Jan 11. Biochemistry. 2012. PMID: 22148590 Free PMC article.
-
GenK-catalyzed C-6' methylation in the biosynthesis of gentamicin: isolation and characterization of a cobalamin-dependent radical SAM enzyme.J Am Chem Soc. 2013 Jun 5;135(22):8093-6. doi: 10.1021/ja312641f. Epub 2013 May 21. J Am Chem Soc. 2013. PMID: 23679096 Free PMC article.
-
Structure and function of phosphonoacetaldehyde dehydrogenase: the missing link in phosphonoacetate formation.Chem Biol. 2014 Jan 16;21(1):125-35. doi: 10.1016/j.chembiol.2013.11.006. Epub 2013 Dec 19. Chem Biol. 2014. PMID: 24361046 Free PMC article.
-
Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.ACS Chem Biol. 2017 Feb 17;12(2):456-463. doi: 10.1021/acschembio.6b00939. Epub 2016 Dec 27. ACS Chem Biol. 2017. PMID: 27977135 Free PMC article.
-
Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology.Curr Opin Chem Biol. 2013 Aug;17(4):580-8. doi: 10.1016/j.cbpa.2013.06.018. Epub 2013 Jul 17. Curr Opin Chem Biol. 2013. PMID: 23870698 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

