YibK is the 2'-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors
- PMID: 20855540
- PMCID: PMC2957053
- DOI: 10.1261/rna.2245910
YibK is the 2'-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors
Abstract
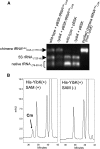
Transfer RNAs are the most densely modified nucleic acid molecules in living cells. In Escherichia coli, more than 30 nucleoside modifications have been characterized, ranging from methylations and pseudouridylations to more complex additions that require multiple enzymatic steps. Most of the modifying enzymes have been identified, although a few notable exceptions include the 2'-O-methyltransferase(s) that methylate the ribose at the nucleotide 34 wobble position in the two leucyl isoacceptors tRNA(Leu)(CmAA) and tRNA(Leu)(cmnm5UmAA). Here, we have used a comparative genomics approach to uncover candidate E. coli genes for the missing enzyme(s). Transfer RNAs from null mutants for candidate genes were analyzed by mass spectrometry and revealed that inactivation of yibK leads to loss of 2'-O-methylation at position 34 in both tRNA(Leu)(CmAA) and tRNA(Leu)(cmnm5UmAA). Loss of YibK methylation reduces the efficiency of codon-wobble base interaction, as demonstrated in an amber suppressor supP system. Inactivation of yibK had no detectable effect on steady-state growth rate, although a distinct disadvantage was noted in multiple-round, mixed-population growth experiments, suggesting that the ability to recover from the stationary phase was impaired. Methylation is restored in vivo by complementing with a recombinant copy of yibK. Despite being one of the smallest characterized α/β knot proteins, YibK independently catalyzes the methyl transfer from S-adenosyl-L-methionine to the 2'-OH of the wobble nucleotide; YibK recognition of this target requires a pyridine at position 34 and N⁶-(isopentenyl)-2-methylthioadenosine at position 37. YibK is one of the last remaining E. coli tRNA modification enzymes to be identified and is now renamed TrmL.
Figures




References
-
- Bishop AC, Xu J, Johnson RC, Schimmel P, de Crecy-Lagard V 2002. Identification of the tRNA-dihydrouridine synthase family. J Biol Chem 277: 25090–25095 - PubMed
-
- Björk GR, Hagervall TG 2005. Transfer RNA modification. In EcoSal—Escherichia coli and Salmonella: Cellular and molecular biology (ed. Böck RCI et al.), Chap. 4.6.2. ASM Press, Washington, DC
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
