INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway
- PMID: 20855601
- PMCID: PMC2951448
- DOI: 10.1073/pnas.1008388107
INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway
Abstract
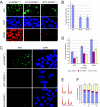
The creation of accessible DNA in the context of chromatin is a key step in many DNA functions. To reveal how ATP-dependent chromatin remodeling activities impact DNA repair, we constructed mammalian genetic models for the INO80 chromatin remodeling complex and investigated the impact of loss of INO80 function on the repair of UV-induced photo lesions. We showed that deletion of two core components of the INO80 complex, INO80 and ARP5, significantly hampered cellular removal of UV-induced photo lesions but had no significant impact on the transcription of nucleotide excision repair (NER) factors. Loss of INO80 abolished the assembly of NER factors, suggesting that prior chromatin relaxation is important for the NER incision process. Ino80 and Arp5 are enriched to UV-damaged DNA in an NER-incision-independent fashion, suggesting that recruitment of the remodeling activity likely takes place during the initial stage of damage recognition. These results demonstrate a critical role of INO80 in creating DNA accessibility for the NER pathway and provide direct evidence that repair of UV lesions and perhaps most bulky adduct lesions requires chromatin reconfiguration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


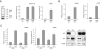
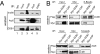
Similar articles
-
The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair.J Cell Biol. 2010 Dec 13;191(6):1061-8. doi: 10.1083/jcb.201006178. Epub 2010 Dec 6. J Cell Biol. 2010. PMID: 21135142 Free PMC article.
-
Assembly of the Arp5 (Actin-related Protein) Subunit Involved in Distinct INO80 Chromatin Remodeling Activities.J Biol Chem. 2015 Oct 16;290(42):25700-9. doi: 10.1074/jbc.M115.674887. Epub 2015 Aug 25. J Biol Chem. 2015. PMID: 26306040 Free PMC article.
-
INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair.Cell. 2004 Dec 17;119(6):767-75. doi: 10.1016/j.cell.2004.11.037. Cell. 2004. PMID: 15607974
-
Assays for chromatin remodeling during nucleotide excision repair in Saccharomyces cerevisiae.Methods. 2009 May;48(1):19-22. doi: 10.1016/j.ymeth.2009.03.017. Epub 2009 Mar 29. Methods. 2009. PMID: 19336254 Free PMC article. Review.
-
Nucleotide excision repair in chromatin: damage removal at the drop of a HAT.DNA Repair (Amst). 2011 Jul 15;10(7):734-42. doi: 10.1016/j.dnarep.2011.04.029. Epub 2011 May 20. DNA Repair (Amst). 2011. PMID: 21600858 Review.
Cited by
-
The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair.Int J Mol Sci. 2012;13(9):11954-11973. doi: 10.3390/ijms130911954. Epub 2012 Sep 20. Int J Mol Sci. 2012. PMID: 23109894 Free PMC article. Review.
-
DDB2 promotes chromatin decondensation at UV-induced DNA damage.J Cell Biol. 2012 Apr 16;197(2):267-81. doi: 10.1083/jcb.201106074. Epub 2012 Apr 9. J Cell Biol. 2012. PMID: 22492724 Free PMC article.
-
Damaged DNA-binding protein down-regulates epigenetic mark H3K56Ac through histone deacetylase 1 and 2.Mutat Res. 2015 Jun;776:16-23. doi: 10.1016/j.mrfmmm.2015.01.005. Epub 2015 Jan 24. Mutat Res. 2015. PMID: 26255936 Free PMC article.
-
BAP1 promotes the repair of UV-induced DNA damage via PARP1-mediated recruitment to damage sites and control of activity and stability.Cell Death Differ. 2022 Dec;29(12):2381-2398. doi: 10.1038/s41418-022-01024-w. Epub 2022 May 30. Cell Death Differ. 2022. PMID: 35637285 Free PMC article.
-
Regulation of DNA Repair Mechanisms: How the Chromatin Environment Regulates the DNA Damage Response.Int J Mol Sci. 2017 Aug 5;18(8):1715. doi: 10.3390/ijms18081715. Int J Mol Sci. 2017. PMID: 28783053 Free PMC article. Review.
References
-
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. - PubMed
-
- Owen-Hughes T, et al. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol Biol. 1999;119:319–331. - PubMed
-
- Peterson CL, Côté J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. - PubMed
-
- Ebbert R, Birkmann A, Schüller H-J. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol Microbiol. 1999;32:741–751. - PubMed
-
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

