Structural Survey of Zinc Containing Proteins and the Development of the Zinc AMBER Force Field (ZAFF)
- PMID: 20856692
- PMCID: PMC2941202
- DOI: 10.1021/ct1002626
Structural Survey of Zinc Containing Proteins and the Development of the Zinc AMBER Force Field (ZAFF)
Abstract
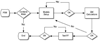
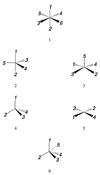
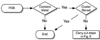
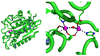
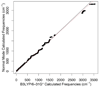
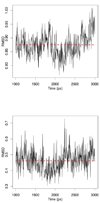
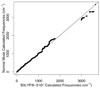
Currently the Protein Data Bank (PDB) contains over 18,000 structures that contain a metal ion including Na, Mg, K, Ca, V, Cr, Mn, Fe, Co, Ni, Cu, Zn, Pd, Ag, Cd, Ir, Pt, Au, and Hg. In general, carrying out classical molecular dynamics (MD) simulations of metalloproteins is a convoluted and time consuming process. Herein, we describe MCPB (Metal Center Parameter Builder), which allows one, to conveniently and rapidly incorporate metal ions using the bonded plus electrostatics model (Hoops et al., J. Am. Chem. Soc. 1991, 113, 8262-8270) into the AMBER Force Field (FF). MCPB was used to develop a Zinc FF, ZAFF, which is compatible with the existing AMBER FFs. The PDB was mined for all Zn containing structures with most being tetrahedrally bound. The most abundant primary shell ligand combinations were extracted and FFs were created. These include Zn bound to CCCC, CCCH, CCHH, CHHH, HHHH, HHHO, HHOO, HOOO, HHHD, and HHDD (O = water and the remaining are 1 letter amino acid codes). Bond and angle force constants and RESP charges were obtained from B3LYP/6-31G* calculations of model structures from the various primary shell combinations. MCPB and ZAFF can be used to create FFs for MD simulations of metalloproteins to study enzyme catalysis, drug design and metalloprotein crystal refinement.
Figures




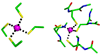

References
-
- Hancock RD. Acc. Chem. Res. 1990;23:253.
-
- Hancock RD. Prog. Inorg. Chem. 1989;37:187.
-
- Hoops SC, Anderson KW, Merz KM., Jr J. Am. Chem. Soc. 1991;113:8262.
-
- Bayly CI, Cieplak P, Cornell WD, Kollman PA. J. Phys. Chem. 1993;97:10269.
-
- Li JB, Zhu TH, Cramer CJ, Truhlar DG. J. Phys. Chem. A. 1998;102:1820.
Grants and funding
LinkOut - more resources
Full Text Sources
