The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.)
- PMID: 20856810
- PMCID: PMC2939885
- DOI: 10.1371/journal.pone.0012762
The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.)
Abstract
Background: Date palm (Phoenix dactylifera L.), a member of Arecaceae family, is one of the three major economically important woody palms--the two other palms being oil palm and coconut tree--and its fruit is a staple food among Middle East and North African nations, as well as many other tropical and subtropical regions. Here we report a complete sequence of the data palm chloroplast (cp) genome based on pyrosequencing.
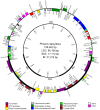
Methodology/principal findings: After extracting 369,022 cp sequencing reads from our whole-genome-shotgun data, we put together an assembly and validated it with intensive PCR-based verification, coupled with PCR product sequencing. The date palm cp genome is 158,462 bp in length and has a typical quadripartite structure of the large (LSC, 86,198 bp) and small single-copy (SSC, 17,712 bp) regions separated by a pair of inverted repeats (IRs, 27,276 bp). Similar to what has been found among most angiosperms, the date palm cp genome harbors 112 unique genes and 19 duplicated fragments in the IR regions. The junctions between LSC/IRs and SSC/IRs show different features of sequence expansion in evolution. We identified 78 SNPs as major intravarietal polymorphisms within the population of a specific cp genome, most of which were located in genes with vital functions. Based on RNA-sequencing data, we also found 18 polycistronic transcription units and three highly expression-biased genes--atpF, trnA-UGC, and rrn23.
Conclusions: Unlike most monocots, date palm has a typical cp genome similar to that of tobacco--with little rearrangement and gene loss or gain. High-throughput sequencing technology facilitates the identification of intravarietal variations in cp genomes among different cultivars. Moreover, transcriptomic analysis of cp genes provides clues for uncovering regulatory mechanisms of transcription and translation in chloroplasts.
Conflict of interest statement
Figures



Similar articles
-
Characterization of the chloroplast genome sequence of oil palm (Elaeis guineensis Jacq.).Gene. 2012 Jun 1;500(2):172-80. doi: 10.1016/j.gene.2012.03.061. Epub 2012 Apr 2. Gene. 2012. PMID: 22487870
-
First chloroplast genomics study of Phoenix dactylifera (var. Naghal and Khanezi): A comparative analysis.PLoS One. 2018 Jul 31;13(7):e0200104. doi: 10.1371/journal.pone.0200104. eCollection 2018. PLoS One. 2018. PMID: 30063732 Free PMC article.
-
Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes.BMC Evol Biol. 2009 Jun 11;9:130. doi: 10.1186/1471-2148-9-130. BMC Evol Biol. 2009. PMID: 19519899 Free PMC article.
-
Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes.Gene. 2012 Oct 15;508(1):96-105. doi: 10.1016/j.gene.2012.07.020. Epub 2012 Jul 25. Gene. 2012. PMID: 22841789
-
An Overview of Date (Phoenix dactylifera) Fruits as an Important Global Food Resource.Foods. 2024 Mar 27;13(7):1024. doi: 10.3390/foods13071024. Foods. 2024. PMID: 38611330 Free PMC article. Review.
Cited by
-
Comparing and phylogenetic analysis chloroplast genome of three Achyranthes species.Sci Rep. 2020 Jul 2;10(1):10818. doi: 10.1038/s41598-020-67679-y. Sci Rep. 2020. PMID: 32616875 Free PMC article.
-
The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza.PLoS One. 2013;8(2):e57607. doi: 10.1371/journal.pone.0057607. Epub 2013 Feb 27. PLoS One. 2013. PMID: 23460883 Free PMC article.
-
The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: insights into the evolution of plant organellar genomes.PLoS One. 2012;7(1):e30531. doi: 10.1371/journal.pone.0030531. Epub 2012 Jan 23. PLoS One. 2012. PMID: 22291979 Free PMC article.
-
CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences.BMC Genomics. 2012 Dec 20;13:715. doi: 10.1186/1471-2164-13-715. BMC Genomics. 2012. PMID: 23256920 Free PMC article.
-
Structure and features of the complete chloroplast genome of Melastoma dodecandrum.Physiol Mol Biol Plants. 2019 Jul;25(4):1043-1054. doi: 10.1007/s12298-019-00651-x. Epub 2019 Mar 12. Physiol Mol Biol Plants. 2019. PMID: 31404219 Free PMC article.
References
-
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–2190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

