Classification of protein kinases on the basis of both kinase and non-kinase regions
- PMID: 20856812
- PMCID: PMC2939887
- DOI: 10.1371/journal.pone.0012460
Classification of protein kinases on the basis of both kinase and non-kinase regions
Abstract
Background: Protein phosphorylation is a generic way to regulate signal transduction pathways in all kingdoms of life. In many organisms, it is achieved by the large family of Ser/Thr/Tyr protein kinases which are traditionally classified into groups and subfamilies on the basis of the amino acid sequence of their catalytic domains. Many protein kinases are multi-domain in nature but the diversity of the accessory domains and their organization are usually not taken into account while classifying kinases into groups or subfamilies.
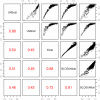
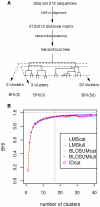
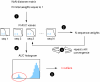
Methodology: Here, we present an approach which considers amino acid sequences of complete gene products, in order to suggest refinements in sets of pre-classified sequences. The strategy is based on alignment-free similarity scores and iterative Area Under the Curve (AUC) computation. Similarity scores are computed by detecting common patterns between two sequences and scoring them using a substitution matrix, with a consistent normalization scheme. This allows us to handle full-length sequences, and implicitly takes into account domain diversity and domain shuffling. We quantitatively validate our approach on a subset of 212 human protein kinases. We then employ it on the complete repertoire of human protein kinases and suggest few qualitative refinements in the subfamily assignment stored in the KinG database, which is based on catalytic domains only. Based on our new measure, we delineate 37 cases of potential hybrid kinases: sequences for which classical classification based entirely on catalytic domains is inconsistent with the full-length similarity scores computed here, which implicitly consider multi-domain nature and regions outside the catalytic kinase domain. We also provide some examples of hybrid kinases of the protozoan parasite Entamoeba histolytica.
Conclusions: The implicit consideration of multi-domain architectures is a valuable inclusion to complement other classification schemes. The proposed algorithm may also be employed to classify other families of enzymes with multi-domain architecture.
Conflict of interest statement
Figures

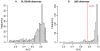


References
-
- Cohen P. Protein kinases–the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. - PubMed
-
- Han E, McGonigal T. Role of focal adhesion kinase in human cancer: a potential target for drug discovery. Anticancer Agents Med Chem. 2007;7:681–684. - PubMed
-
- Hardie D. AMP-activated protein kinase as a drug target. Annu Rev Pharmacol Toxicol. 2007;47:185–210. - PubMed
-
- Hunter T, Plowman G. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

