The unfolded protein response is not necessary for the G1/S transition, but it is required for chromosome maintenance in Saccharomyces cerevisiae
- PMID: 20856872
- PMCID: PMC2939067
- DOI: 10.1371/journal.pone.0012732
The unfolded protein response is not necessary for the G1/S transition, but it is required for chromosome maintenance in Saccharomyces cerevisiae
Abstract
Background: The unfolded protein response (UPR) is a eukaryotic signaling pathway, from the endoplasmic reticulum (ER) to the nucleus. Protein misfolding in the ER triggers the UPR. Accumulating evidence links the UPR in diverse aspects of cellular homeostasis. The UPR responds to the overall protein synthesis capacity and metabolic fluxes of the cell. Because the coupling of metabolism with cell division governs when cells start dividing, here we examined the role of UPR signaling in the timing of initiation of cell division and cell cycle progression, in the yeast Saccharomyces cerevisiae.
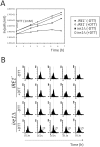
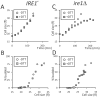
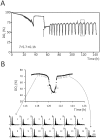
Methodology/principal findings: We report that cells lacking the ER-resident stress sensor Ire1p, which cannot trigger the UPR, nonetheless completed the G1/S transition on time. Furthermore, loss of UPR signaling neither affected the nutrient and growth rate dependence of the G1/S transition, nor the metabolic oscillations that yeast cells display in defined steady-state conditions. Remarkably, however, loss of UPR signaling led to hypersensitivity to genotoxic stress and a ten-fold increase in chromosome loss.
Conclusions/significance: Taken together, our results strongly suggest that UPR signaling is not necessary for the normal coupling of metabolism with cell division, but it has a role in genome maintenance. These results add to previous work that linked the UPR with cytokinesis in yeast. UPR signaling is conserved in all eukaryotes, and it malfunctions in a variety of diseases, including cancer. Therefore, our findings may be relevant to other systems, including humans.
Conflict of interest statement
Figures




Similar articles
-
Fast-Growing Saccharomyces cerevisiae Cells with a Constitutive Unfolded Protein Response and Their Potential for Lipidic Molecule Production.Appl Environ Microbiol. 2022 Nov 8;88(21):e0108322. doi: 10.1128/aem.01083-22. Epub 2022 Oct 18. Appl Environ Microbiol. 2022. PMID: 36255243 Free PMC article.
-
Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae.Proc Natl Acad Sci U S A. 2017 Mar 21;114(12):E2411-E2419. doi: 10.1073/pnas.1612422114. Epub 2017 Mar 6. Proc Natl Acad Sci U S A. 2017. PMID: 28265091 Free PMC article.
-
4-Phenylbutyrate suppresses the unfolded protein response without restoring protein folding in Saccharomyces cerevisiae.FEMS Yeast Res. 2018 Mar 1;18(2). doi: 10.1093/femsyr/foy016. FEMS Yeast Res. 2018. PMID: 29452364
-
The cellular response to protein misfolding in the endoplasmic reticulum.Gene Expr. 1999;7(4-6):293-300. Gene Expr. 1999. PMID: 10440230 Free PMC article. Review.
-
Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae.Int J Mol Sci. 2019 Jun 12;20(12):2860. doi: 10.3390/ijms20122860. Int J Mol Sci. 2019. PMID: 31212749 Free PMC article. Review.
Cited by
-
A systematic analysis of cell cycle regulators in yeast reveals that most factors act independently of cell size to control initiation of division.PLoS Genet. 2012;8(3):e1002590. doi: 10.1371/journal.pgen.1002590. Epub 2012 Mar 15. PLoS Genet. 2012. PMID: 22438835 Free PMC article.
-
Scaling of G1 Duration with Population Doubling Time by a Cyclin in Saccharomyces cerevisiae.Genetics. 2018 Nov;210(3):895-906. doi: 10.1534/genetics.118.301507. Epub 2018 Aug 27. Genetics. 2018. PMID: 30150288 Free PMC article.
-
Signaling networks converge on TORC1-SREBP activity to promote endoplasmic reticulum homeostasis.PLoS One. 2014 Jul 9;9(7):e101164. doi: 10.1371/journal.pone.0101164. eCollection 2014. PLoS One. 2014. PMID: 25007267 Free PMC article.
-
A nonenzymatic dependency on inositol-requiring enzyme 1 controls cancer cell cycle progression and tumor growth.PLoS Biol. 2025 Apr 10;23(4):e3003086. doi: 10.1371/journal.pbio.3003086. eCollection 2025 Apr. PLoS Biol. 2025. PMID: 40208872 Free PMC article.
-
PMT1 deficiency enhances basal UPR activity and extends replicative lifespan of Saccharomyces cerevisiae.Age (Dordr). 2015 Jun;37(3):9788. doi: 10.1007/s11357-015-9788-7. Epub 2015 May 4. Age (Dordr). 2015. PMID: 25936926 Free PMC article.
References
-
- Patil C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol. 2001;13:349–355. - PubMed
-
- Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. - PubMed
-
- Zhao L, Ackerman SL. Endoplasmic reticulum stress in health and disease. Curr Opin Cell Biol. 2006;18:444–452. - PubMed
-
- Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, et al. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol. 2002;3:411–421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

