Active site conformational dynamics in human uridine phosphorylase 1
- PMID: 20856879
- PMCID: PMC2939078
- DOI: 10.1371/journal.pone.0012741
Active site conformational dynamics in human uridine phosphorylase 1
Abstract
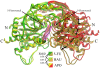
Uridine phosphorylase (UPP) is a central enzyme in the pyrimidine salvage pathway, catalyzing the reversible phosphorolysis of uridine to uracil and ribose-1-phosphate. Human UPP activity has been a focus of cancer research due to its role in activating fluoropyrimidine nucleoside chemotherapeutic agents such as 5-fluorouracil (5-FU) and capecitabine. Additionally, specific molecular inhibitors of this enzyme have been found to raise endogenous uridine concentrations, which can produce a cytoprotective effect on normal tissues exposed to these drugs. Here we report the structure of hUPP1 bound to 5-FU at 2.3 A resolution. Analysis of this structure reveals new insights as to the conformational motions the enzyme undergoes in the course of substrate binding and catalysis. The dimeric enzyme is capable of a large hinge motion between its two domains, facilitating ligand exchange and explaining observed cooperativity between the two active sites in binding phosphate-bearing substrates. Further, a loop toward the back end of the uracil binding pocket is shown to flexibly adjust to the varying chemistry of different compounds through an "induced-fit" association mechanism that was not observed in earlier hUPP1 structures. The details surrounding these dynamic aspects of hUPP1 structure and function provide unexplored avenues to develop novel inhibitors of this protein with improved specificity and increased affinity. Given the recent emergence of new roles for uridine as a neuron protective compound in ischemia and degenerative diseases, such as Alzheimer's and Parkinson's, inhibitors of hUPP1 with greater efficacy, which are able to boost cellular uridine levels without adverse side-effects, may have a wide range of therapeutic applications.
Conflict of interest statement
Figures

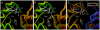

Similar articles
-
A novel structural mechanism for redox regulation of uridine phosphorylase 2 activity.J Struct Biol. 2011 Nov;176(2):229-37. doi: 10.1016/j.jsb.2011.08.002. Epub 2011 Aug 10. J Struct Biol. 2011. PMID: 21855639 Free PMC article.
-
Implications of the structure of human uridine phosphorylase 1 on the development of novel inhibitors for improving the therapeutic window of fluoropyrimidine chemotherapy.BMC Struct Biol. 2009 Mar 16;9:14. doi: 10.1186/1472-6807-9-14. BMC Struct Biol. 2009. PMID: 19291308 Free PMC article.
-
Crystal structures of Escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium.J Mol Biol. 2004 Mar 19;337(2):337-54. doi: 10.1016/j.jmb.2004.01.039. J Mol Biol. 2004. PMID: 15003451
-
Uridine phosophorylase: an important enzyme in pyrimidine metabolism and fluoropyrimidine activation.Drugs Today (Barc). 2004 May;40(5):431-43. doi: 10.1358/dot.2004.40.5.850491. Drugs Today (Barc). 2004. PMID: 15319798 Review.
-
Structural analyses reveal two distinct families of nucleoside phosphorylases.Biochem J. 2002 Jan 1;361(Pt 1):1-25. doi: 10.1042/0264-6021:3610001. Biochem J. 2002. PMID: 11743878 Free PMC article. Review.
Cited by
-
A novel structural mechanism for redox regulation of uridine phosphorylase 2 activity.J Struct Biol. 2011 Nov;176(2):229-37. doi: 10.1016/j.jsb.2011.08.002. Epub 2011 Aug 10. J Struct Biol. 2011. PMID: 21855639 Free PMC article.
-
Structural and catalytic analysis of two diverse uridine phosphorylases in Phytophthora capsici.Sci Rep. 2020 Jun 3;10(1):9051. doi: 10.1038/s41598-020-65935-9. Sci Rep. 2020. PMID: 32493959 Free PMC article.
-
Metabolites modulate the functional state of human uridine phosphorylase I.Protein Sci. 2020 Nov;29(11):2189-2200. doi: 10.1002/pro.3939. Epub 2020 Sep 28. Protein Sci. 2020. PMID: 32864839 Free PMC article.
-
Uridine phosphorylase 1 associates to biological and clinical significance in thyroid carcinoma cell lines.J Cell Mol Med. 2019 Nov;23(11):7438-7448. doi: 10.1111/jcmm.14612. Epub 2019 Sep 9. J Cell Mol Med. 2019. PMID: 31496029 Free PMC article.
-
Pathway analysis for drug repositioning based on public database mining.J Chem Inf Model. 2014 Feb 24;54(2):407-18. doi: 10.1021/ci4005354. Epub 2014 Feb 5. J Chem Inf Model. 2014. PMID: 24460210 Free PMC article.
References
-
- Cao D, Pizzorno G. Uridine phosophorylase: an important enzyme in pyrimidine metabolism and fluoropyrimidine activation. Drugs Today (Barc) 2004;40:431–443. - PubMed
-
- Cappiello M, Mascia L, Scolozzi C, Giorgelli F, Ipata PL. In vitro assessment of salvage pathways for pyrimidine bases in rat liver and brain. Biochim Biophys Acta. 1998;1425:273–281. - PubMed
-
- Tozzi MG, Camici M, Mascia L, Sgarrella F, Ipata PL. Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 2006;273:1089–1101. - PubMed
-
- Watanabe S, Uchida T. Cloning and expression of human uridine phosphorylase. Biochem Biophys Res Commun. 1995;216:265–272. - PubMed
-
- Johansson M. Identification of a novel human uridine phosphorylase. Biochem Biophys Res Commun. 2003;307:41–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

