In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM)
- PMID: 20856929
- PMCID: PMC2938336
- DOI: 10.1371/journal.pone.0012680
In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM)
Abstract
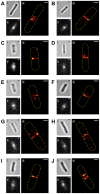
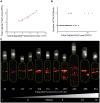
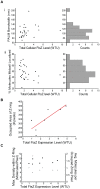
The FtsZ protein, a tubulin-like GTPase, plays a pivotal role in prokaryotic cell division. In vivo it localizes to the midcell and assembles into a ring-like structure-the Z-ring. The Z-ring serves as an essential scaffold to recruit all other division proteins and generates contractile force for cytokinesis, but its supramolecular structure remains unknown. Electron microscopy (EM) has been unsuccessful in detecting the Z-ring due to the dense cytoplasm of bacterial cells, and conventional fluorescence light microscopy (FLM) has only provided images with limited spatial resolution (200-300 nm) due to the diffraction of light. Hence, given the small sizes of bacteria cells, identifying the in vivo structure of the Z-ring presents a substantial challenge. Here, we used photoactivated localization microscopy (PALM), a single molecule-based super-resolution imaging technique, to characterize the in vivo structure of the Z-ring in E. coli. We achieved a spatial resolution of ∼35 nm and discovered that in addition to the expected ring-like conformation, the Z-ring of E. coli adopts a novel compressed helical conformation with variable helical length and pitch. We measured the thickness of the Z-ring to be ∼110 nm and the packing density of FtsZ molecules inside the Z-ring to be greater than what is expected for a single-layered flat ribbon configuration. Our results strongly suggest that the Z-ring is composed of a loose bundle of FtsZ protofilaments that randomly overlap with each other in both longitudinal and radial directions of the cell. Our results provide significant insight into the spatial organization of the Z-ring and open the door for further investigations of structure-function relationships and cell cycle-dependent regulation of the Z-ring.
Conflict of interest statement
Figures





References
-
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. - PubMed
-
- Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. - PubMed
-
- RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

