Human common salivary protein 1 (CSP-1) promotes binding of Streptococcus mutans to experimental salivary pellicle and glucans formed on hydroxyapatite surface
- PMID: 20858015
- PMCID: PMC2997150
- DOI: 10.1021/pr100786y
Human common salivary protein 1 (CSP-1) promotes binding of Streptococcus mutans to experimental salivary pellicle and glucans formed on hydroxyapatite surface
Abstract
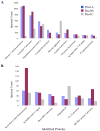
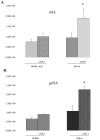
The saliva proteome includes host defense factors and specific bacterial-binding proteins that modulate microbial growth and colonization of the tooth surface in the oral cavity. A multidimensional mass spectrometry approach identified the major host-derived salivary proteins that interacted with Streptococcus mutans (strain UA159), the primary microorganism associated with the pathogenesis of dental caries. Two abundant host proteins were found to tightly bind to S. mutans cells, common salivary protein-1 (CSP-1) and deleted in malignant brain tumor 1 (DMBT1, also known as salivary agglutinin or gp340). In contrast to gp340, limited functional information is available on CSP-1. The sequence of CSP-1 shares 38.1% similarity with rat CSP-1. Recombinant CSP-1 (rCSP-1) protein did not cause aggregation of S. mutans cells and was devoid of any significant biocidal activity (2.5 to 10 μg/mL). However, S. mutans cells exposed to rCSP-1 (10 μg/mL) in saliva displayed enhanced adherence to experimental salivary pellicle and to glucans in the pellicle formed on hydroxyapatite surfaces. Thus, our data demonstrate that the host salivary protein CSP-1 binds to S. mutans cells and may influence the initial colonization of this pathogenic bacterium onto the tooth surface.
Figures


References
-
- Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J Dent Res. 2007;86:680–693. - PubMed
-
- Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, Delahunty CM, Denny T, Dunsmore J, Faull KF, Gilligan J, Gonzalez-Begne M, Halgand F, Hall SC, Han X, Henson B, Hewel J, Hu S, Jeffrey S, Jiang J, Loo JA, Ogorzalek Loo RR, Malamud D, Melvin JE, Miroshnychenko O, Navazesh M, Niles R, Park SK, Prakobphol A, Ramachandran P, Richert M, Robinson S, Sondej M, Souda P, Sullivan MA, Takashima J, Than S, Wang J, Whitelegge JP, Witkowska HE, Wolinsky L, Xie Y, Xu T, Yu W, Ytterberg J, Wong DT, Yates JRIII, Fisher SJ. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994–2006. - PMC - PubMed
-
- Douglas CWI. The binding of human salivary a-amylase by oral strains of streptococcal bacteria. Arch Oral Biol. 1983;28:567–573. - PubMed
-
- Marsh PD. Host defense and microbial homeostasis: role of microbial interaction. J Dent Res. 1989;68:1567–1575.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

