MicroRNA roles in beta-catenin pathway
- PMID: 20858269
- PMCID: PMC2955614
- DOI: 10.1186/1476-4598-9-252
MicroRNA roles in beta-catenin pathway
Abstract
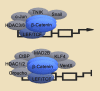
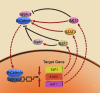
β-catenin, a key factor in the Wnt signaling pathway, has essential functions in the regulation of cell growth and differentiation. Aberrant β-catenin signaling has been linked to various disease pathologies, including an important role in tumorigenesis. Here, we review the regulation of the Wnt signaling pathway as it relates to β-catenin signaling in tumorigenesis, with particular focus on the role of microRNAs. Finally, we discuss the potential of β-catenin targeted therapeutics for cancer treatment.
Figures



Similar articles
-
MicroRNA-200a suppresses the Wnt/β-catenin signaling pathway by interacting with β-catenin.Int J Oncol. 2012 Apr;40(4):1162-70. doi: 10.3892/ijo.2011.1322. Epub 2011 Dec 30. Int J Oncol. 2012. PMID: 22211245 Free PMC article.
-
Distinctive microRNA signature associated of neoplasms with the Wnt/β-catenin signaling pathway.Cell Signal. 2013 Dec;25(12):2805-11. doi: 10.1016/j.cellsig.2013.09.006. Epub 2013 Sep 13. Cell Signal. 2013. PMID: 24041653 Review.
-
Role of regulatory miRNAs of the Wnt/ β-catenin signaling pathway in tumorigenesis of breast cancer.Gene. 2020 Sep 5;754:144892. doi: 10.1016/j.gene.2020.144892. Epub 2020 Jun 10. Gene. 2020. PMID: 32534060 Review.
-
Convergence between Wnt-β-catenin and EGFR signaling in cancer.Mol Cancer. 2010 Sep 9;9:236. doi: 10.1186/1476-4598-9-236. Mol Cancer. 2010. PMID: 20828404 Free PMC article. Review.
-
MicroRNAs target the Wnt/β‑catenin signaling pathway to regulate epithelial‑mesenchymal transition in cancer (Review).Oncol Rep. 2020 Oct;44(4):1299-1313. doi: 10.3892/or.2020.7703. Epub 2020 Jul 23. Oncol Rep. 2020. PMID: 32700744 Free PMC article. Review.
Cited by
-
MicroRNA-142-3p Negatively Regulates Canonical Wnt Signaling Pathway.PLoS One. 2016 Jun 27;11(6):e0158432. doi: 10.1371/journal.pone.0158432. eCollection 2016. PLoS One. 2016. PMID: 27348426 Free PMC article.
-
MicroRNA-200a suppresses the Wnt/β-catenin signaling pathway by interacting with β-catenin.Int J Oncol. 2012 Apr;40(4):1162-70. doi: 10.3892/ijo.2011.1322. Epub 2011 Dec 30. Int J Oncol. 2012. PMID: 22211245 Free PMC article.
-
The effect of baicalein on Wnt/β-catenin pathway and miR-25 expression in Saos-2 osteosarcoma cell line.Turk J Med Sci. 2020 Jun 23;50(44):1168-1179. doi: 10.3906/sag-2001-161. Turk J Med Sci. 2020. PMID: 32283909 Free PMC article.
-
The role of MiRNA-21 in gliomas: Hope for a novel therapeutic intervention?Toxicol Rep. 2020 Nov 6;7:1514-1530. doi: 10.1016/j.toxrep.2020.11.001. eCollection 2020. Toxicol Rep. 2020. PMID: 33251119 Free PMC article.
-
Epigenetics of Meningiomas.Biomed Res Int. 2015;2015:532451. doi: 10.1155/2015/532451. Epub 2015 May 25. Biomed Res Int. 2015. PMID: 26101774 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

