High cell density and latent membrane protein 1 expression induce cleavage of the mixed lineage leukemia gene at 11q23 in nasopharyngeal carcinoma cell line
- PMID: 20858288
- PMCID: PMC2954915
- DOI: 10.1186/1423-0127-17-77
High cell density and latent membrane protein 1 expression induce cleavage of the mixed lineage leukemia gene at 11q23 in nasopharyngeal carcinoma cell line
Abstract
Background: Nasopharyngeal carcinoma (NPC) is commonly found in Southern China and South East Asia. Epstein-Barr virus (EBV) infection is well associated with NPC and has been implicated in its pathogenesis. Moreover, various chromosome rearrangements were reported in NPC. However, the underlying mechanism of chromosome rearrangement remains unclear. Furthermore, the relationship between EBV and chromosome rearrangement with respect to the pathogenesis of NPC has not been established. We hypothesize that during virus- or stress-induced apoptosis, chromosomes are initially cleaved at the base of the chromatin loop domain structure. Upon DNA repair, cell may survive with rearranged chromosomes.
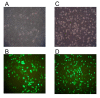
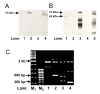
Methods: In this study, cells were seeded at various densities to induce apoptosis. Genomic DNA extracted was processed for Southern hybridization. In order to investigate the role of EBV, especially the latent membrane protein 1 (LMP1), LMP1 gene was overexpressed in NPC cells and chromosome breaks were analyzed by inverse polymerase chain (IPCR) reaction.
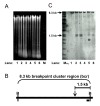
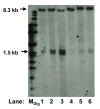
Results: Southern analysis revealed that high cell density resulted in cleavage of the mixed lineage leukemia (MLL) gene within the breakpoint cluster region (bcr). This high cell density-induced cleavage was significantly reduced by caspase inhibitor, Z-DEVD-FMK. Similarly, IPCR analysis showed that LMP1 expression enhanced cleavage of the MLL bcr. Breakpoint analysis revealed that these breaks occurred within the matrix attachment region/scaffold attachment region (MAR/SAR).
Conclusions: Since MLL locates at 11q23, a common deletion site in NPC, our results suggest a possibility of stress- or virus-induced apoptosis in the initiation of chromosome rearrangements at 11q23. The breakpoint analysis results also support the role of chromatin structure in defining the site of chromosome rearrangement.
Figures
References
-
- Fandi A, Altun M, Azli N, Armand JP, Cvitkovic E. Nasopharyngeal cancer: epidemiology, staging, and treatment. Semin Oncol. 1994;21:382–397. - PubMed
-
- Raab-Traub N. Epstein-Barr virus and nasopharyngeal carcinoma. Semin Cancer Biol. 1992;3:297–307. - PubMed
-
- Lin JC, Liao SK, Lee EH, Hung MS, Sayion Y, Chen HC, Kang CC, Huang LS, Cherng JM. Molecular events associated with epithelial to mesenchymal transition of nasopharyngeal carcinoma cells in the absence of Epstein-Barr virus genome. J Biomed Sci. 2009;16:105. doi: 10.1186/1423-0127-16-105. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

