Water under the BAR
- PMID: 20858422
- PMCID: PMC2941004
- DOI: 10.1016/j.bpj.2010.06.074
Water under the BAR
Abstract
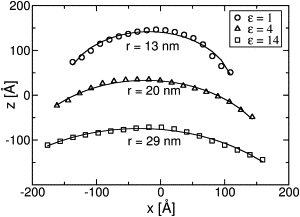
Many cellular processes require the generation of highly curved regions of cell membranes by interfacial membrane proteins. A number of such proteins are now known, and several mechanisms of curvature generation have been suggested, but so far a quantitative understanding of the importance of the various potential mechanisms remains elusive. Following previous theoretical work, we consider the electrostatic attraction that underlies the scaffold mechanism of membrane bending in the context of the N-BAR domain of amphiphysin. Analysis of atomistic molecular dynamics simulations reveals considerable water between the membrane and the positively charged concave face of the BAR, even when it is tightly bound to highly curved membranes. This results in significant screening of electrostatic interactions, suggesting that electrostatic attraction is not the main driving force behind curvature sensing, supporting recent experimental work. These results also emphasize the need for care when building coarse-grained models of protein-membrane interactions. These results are emphasized by simulations of oligomerized amphiphysin N-BARs at the atomistic and coarse-grained level. In the coarse-grained simulations, we find a strong dependence of the induced curvature on the dielectric screening.
Copyright © 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Direct observation of Bin/amphiphysin/Rvs (BAR) domain-induced membrane curvature by means of molecular dynamics simulations.Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):15068-72. doi: 10.1073/pnas.0603917103. Epub 2006 Sep 28. Proc Natl Acad Sci U S A. 2006. PMID: 17008407 Free PMC article.
-
Roles of PIP2 in the membrane binding of MIM I-BAR: insights from molecular dynamics simulations.FEBS Lett. 2018 Aug;592(15):2533-2542. doi: 10.1002/1873-3468.13186. Epub 2018 Jul 26. FEBS Lett. 2018. PMID: 29995324
-
Modeling membrane deformations and lipid demixing upon protein-membrane interaction: the BAR dimer adsorption.Biophys J. 2009 Sep 16;97(6):1626-35. doi: 10.1016/j.bpj.2009.07.006. Biophys J. 2009. PMID: 19751667 Free PMC article.
-
Membrane curvature: how BAR domains bend bilayers.Curr Biol. 2004 Mar 23;14(6):R250-2. doi: 10.1016/j.cub.2004.02.060. Curr Biol. 2004. PMID: 15043839 Review.
-
Attachment of rod-like (BAR) proteins and membrane shape.Mini Rev Med Chem. 2011 Apr;11(4):272-82. doi: 10.2174/138955711795305353. Mini Rev Med Chem. 2011. PMID: 21428902 Free PMC article. Review.
Cited by
-
α-Synuclein-induced membrane remodeling is driven by binding affinity, partition depth, and interleaflet order asymmetry.J Am Chem Soc. 2014 Jul 16;136(28):9962-72. doi: 10.1021/ja5016958. Epub 2014 Jul 7. J Am Chem Soc. 2014. PMID: 24960410 Free PMC article.
-
Salt Bridge Formation between the I-BAR Domain and Lipids Increases Lipid Density and Membrane Curvature.Sci Rep. 2017 Jul 28;7(1):6808. doi: 10.1038/s41598-017-06334-5. Sci Rep. 2017. PMID: 28754893 Free PMC article.
-
Solvent-Free, Highly Coarse-Grained Models for Charged Lipid Systems.J Chem Theory Comput. 2014 Oct 14;10(10):4730-4744. doi: 10.1021/ct500474a. Epub 2014 Sep 10. J Chem Theory Comput. 2014. PMID: 25328498 Free PMC article.
-
Organizing membrane-curving proteins: the emerging dynamical picture.Curr Opin Struct Biol. 2018 Aug;51:99-105. doi: 10.1016/j.sbi.2018.03.018. Epub 2018 Mar 30. Curr Opin Struct Biol. 2018. PMID: 29609179 Free PMC article. Review.
-
Application of a free-energy-landscape approach to study tension-dependent bilayer tubulation mediated by curvature-inducing proteins.Phys Rev E Stat Nonlin Soft Matter Phys. 2015 Oct;92(4):042715. doi: 10.1103/PhysRevE.92.042715. Epub 2015 Oct 29. Phys Rev E Stat Nonlin Soft Matter Phys. 2015. PMID: 26565280 Free PMC article.
References
-
- McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. - PubMed
-
- Zimmerberg J., Kozlov M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. - PubMed
-
- Slepnev V.I., De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat. Rev. Neurosci. 2000;1:161–172. - PubMed
-
- Zimmerberg J., McLaughlin S. Membrane curvature: how BAR domains bend bilayers. Curr. Biol. 2004;14:R250–R252. - PubMed
-
- Peter B.J., Kent H.M., McMahon H.T. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

