Toward a consensus view of duplex RNA flexibility
- PMID: 20858433
- PMCID: PMC2941024
- DOI: 10.1016/j.bpj.2010.06.061
Toward a consensus view of duplex RNA flexibility
Abstract
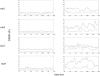
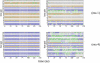
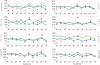
The structure and flexibility of the RNA duplex has been studied using extended molecular dynamics simulations on four diverse 18-mer oligonucleotides designed to contain many copies of the 10 unique dinucleotide steps in different sequence environments. Simulations were performed using the two most popular force fields for nucleic acids simulations (AMBER and CHARMM) in their latest versions, trying to arrive to a consensus picture of the RNA flexibility. Contrary to what was found for DNA duplex (DNA(2)), no clear convergence is found for the RNA duplex (RNA(2)), but one of the force field seems to agree better with experimental data. MD simulations performed with this force field were used to fully characterize, for the first time to our knowledge, the sequence-dependent elastic properties of RNA duplexes at different levels of resolutions. The flexibility pattern of RNA(2) shows similarities with DNA(2), but also surprising differences, which help us to understand the different biological functions of both molecules. A full mesoscopic model of RNA duplex at different resolution levels is derived to be used for genome-wide description of the flexibility of double-helical fragments of RNA.
Copyright © 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Sequence-dependent mechanical properties of double-stranded RNA.Nanoscale. 2019 Nov 28;11(44):21471-21478. doi: 10.1039/c9nr07516j. Epub 2019 Nov 5. Nanoscale. 2019. PMID: 31686065
-
Structural variations of single and tandem mismatches in RNA duplexes: a joint MD simulation and crystal structure database analysis.J Phys Chem B. 2012 Oct 4;116(39):11845-56. doi: 10.1021/jp305628v. Epub 2012 Sep 20. J Phys Chem B. 2012. PMID: 22953716
-
Structural Flexibility of DNA-RNA Hybrid Duplex: Stretching and Twist-Stretch Coupling.Biophys J. 2019 Jul 9;117(1):74-86. doi: 10.1016/j.bpj.2019.05.018. Epub 2019 May 23. Biophys J. 2019. PMID: 31164196 Free PMC article.
-
A molecular view of DNA flexibility.Q Rev Biophys. 2021 Jul 6;54:e8. doi: 10.1017/S0033583521000068. Q Rev Biophys. 2021. PMID: 34225835 Review.
-
Frontiers in molecular dynamics simulations of DNA.Acc Chem Res. 2012 Feb 21;45(2):196-205. doi: 10.1021/ar2001217. Epub 2011 Aug 10. Acc Chem Res. 2012. PMID: 21830782 Review.
Cited by
-
Understanding the Relative Flexibility of RNA and DNA Duplexes: Stretching and Twist-Stretch Coupling.Biophys J. 2017 Mar 28;112(6):1094-1104. doi: 10.1016/j.bpj.2017.02.022. Biophys J. 2017. PMID: 28355538 Free PMC article.
-
Mesoscopic model parametrization of hydrogen bonds and stacking interactions of RNA from melting temperatures.Nucleic Acids Res. 2013 Jan 7;41(1):e30. doi: 10.1093/nar/gks964. Epub 2012 Oct 18. Nucleic Acids Res. 2013. PMID: 23087379 Free PMC article.
-
Isosteric and nonisosteric base pairs in RNA motifs: molecular dynamics and bioinformatics study of the sarcin-ricin internal loop.J Phys Chem B. 2013 Nov 21;117(46):14302-19. doi: 10.1021/jp408530w. Epub 2013 Nov 12. J Phys Chem B. 2013. PMID: 24144333 Free PMC article.
-
Ribonuclease III mechanisms of double-stranded RNA cleavage.Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):31-48. doi: 10.1002/wrna.1195. Epub 2013 Sep 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24124076 Free PMC article. Review.
-
Regulation of the activity of the promoter of RNA-induced silencing, C3PO.Protein Sci. 2017 Sep;26(9):1807-1818. doi: 10.1002/pro.3219. Epub 2017 Jul 17. Protein Sci. 2017. PMID: 28714243 Free PMC article.
References
-
- Brenner S., Jacob F., Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961;190:576–581. - PubMed
-
- Joyce G.F. RNA evolution and the origins of life. Nature. 1989;338:217–224. - PubMed
-
- Cech T.R., Damberger S.H., Gutell R.R. Representation of the secondary and tertiary structure of group I introns. Nat. Struct. Biol. 1994;1:273–280. - PubMed
-
- Cech T.R., Bass B.L. Biological catalysis by RNA. Annu. Rev. Biochem. 1986;55:599–629. - PubMed
-
- Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

