Effect of single-strand break on branch migration and folding dynamics of Holliday junctions
- PMID: 20858437
- PMCID: PMC2941029
- DOI: 10.1016/j.bpj.2010.07.011
Effect of single-strand break on branch migration and folding dynamics of Holliday junctions
Abstract
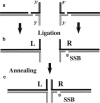
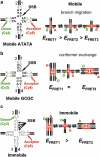
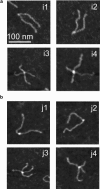
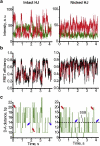
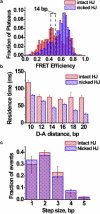
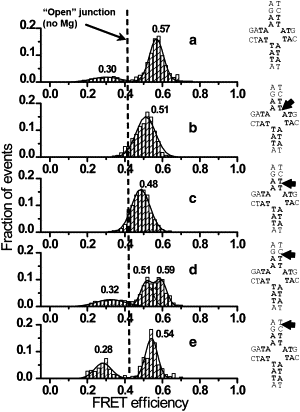
The Holliday junction (HJ), or four-way junction, is a central intermediate state of DNA for homologous genetic recombination and other genetic processes such as replication and repair. Branch migration is the process by which the exchange of homologous DNA regions occurs, and it can be spontaneous or driven by proteins. Unfolding of the HJ is required for branch migration. Our previous single-molecule fluorescence studies led to a model according to which branch migration is a stepwise process consisting of consecutive migration and folding steps. Folding of the HJ in one of the folded conformations terminates the branch migration phase. At the same time, in the unfolded state HJ rapidly migrates over entire homology region of the HJ in one hop. This process can be affected by irregularities in the DNA double helical structure, so mismatches almost terminate a spontaneous branch migration. Single-stranded breaks or nicks are the most ubiquitous defects in the DNA helix; however, to date, their effect on the HJ branch migration has not been studied. In addition, although nicked HJs are specific substrates for a number of enzymes involved in DNA recombination and repair, the role of this substrate specificity remains unclear. Our main goal in this work was to study the effect of nicks on the efficiency of HJ branch migration and the dynamics of the HJ. To accomplish this goal, we applied two single-molecule methods: atomic force microscopy and fluorescence resonance energy transfer. The atomic force microscopy data show that the nick does not prevent branch migration, but it does decrease the probability that the HJ will pass the DNA lesion. The single-molecule fluorescence resonance energy transfer approaches were instrumental in detailing the effects of nicks. These studies reveal a dramatic change of the HJ dynamics. The nick changes the structure and conformational dynamics of the junctions, leading to conformations with geometries that are different from those for the intact HJ. On the basis of these data, we propose a model of branch migration in which the propensity of the junction to unfold decreases the lifetimes of folded states, thereby increasing the frequency of junction fluctuations between the folded states.
Copyright © 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Structure, dynamics, and branch migration of a DNA Holliday junction: a single-molecule fluorescence and modeling study.Biophys J. 2008 Nov 1;95(9):4372-83. doi: 10.1529/biophysj.108.135103. Epub 2008 Jul 25. Biophys J. 2008. PMID: 18658216 Free PMC article.
-
Holliday junction dynamics and branch migration: single-molecule analysis.Proc Natl Acad Sci U S A. 2005 Jun 7;102(23):8186-91. doi: 10.1073/pnas.0407210102. Epub 2005 May 25. Proc Natl Acad Sci U S A. 2005. PMID: 15917329 Free PMC article.
-
Rad54 protein promotes branch migration of Holliday junctions.Nature. 2006 Aug 3;442(7102):590-3. doi: 10.1038/nature04889. Epub 2006 Jul 2. Nature. 2006. PMID: 16862129
-
Homologous Recombination under the Single-Molecule Fluorescence Microscope.Int J Mol Sci. 2019 Dec 3;20(23):6102. doi: 10.3390/ijms20236102. Int J Mol Sci. 2019. PMID: 31816946 Free PMC article. Review.
-
GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution.Genes Dev. 2010 Mar 15;24(6):521-36. doi: 10.1101/gad.1903510. Epub 2010 Mar 4. Genes Dev. 2010. PMID: 20203129 Free PMC article. Review.
Cited by
-
Implications of Metastable Nicks and Nicked Holliday Junctions in Processing Joint Molecules in Mitosis and Meiosis.Genes (Basel). 2020 Dec 12;11(12):1498. doi: 10.3390/genes11121498. Genes (Basel). 2020. PMID: 33322845 Free PMC article.
-
Active Control of Repetitive Structural Transitions between Replication Forks and Holliday Junctions by Werner Syndrome Helicase.Structure. 2016 Aug 2;24(8):1292-1300. doi: 10.1016/j.str.2016.06.004. Epub 2016 Jul 14. Structure. 2016. PMID: 27427477 Free PMC article.
-
Junction resolving enzymes use multivalency to keep the Holliday junction dynamic.Nat Chem Biol. 2019 Mar;15(3):269-275. doi: 10.1038/s41589-018-0209-y. Epub 2019 Jan 21. Nat Chem Biol. 2019. PMID: 30664685 Free PMC article.
References
-
- Holliday R. A mechanism for gene conversion in fungi. Genet. Res. Camb. 1964;5:282–304. - PubMed
-
- Leach D.R.F. Blackwell Science; Oxford, UK/Cambridge, MA: 1996. Genetic Recombination.
-
- Postow L., Ullsperger C., Cozzarelli N.R. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 2001;276:2790–2796. - PubMed
-
- Cromie G.A., Connelly J.C., Leach D.R. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell. 2001;8:1163–1174. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

