Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart
- PMID: 20858598
- PMCID: PMC2972695
- DOI: 10.1093/hmg/ddq393
Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart
Abstract
Heart formation requires a highly balanced network of transcriptional activation of genes. The homeodomain transcription factor, Shox2, is essential for the formation of the sinoatrial valves and for the development of the pacemaking system. The elucidation of molecular mechanisms underlying the development of pacemaker tissue has gained clinical interest as defects in its patterning can be related to atrial arrhythmias. We have analyzed putative targets of Shox2 and identified the Bmp4 gene as a direct target. Shox2 interacts directly with the Bmp4 promoter in chromatin immunoprecipitation assays and activates transcription in luciferase-reporter assays. In addition, ectopic expression of Shox2 in Xenopus embryos stimulates transcription of the Bmp4 gene, and silencing of Shox2 in cardiomyocytes leads to a reduction in the expression of Bmp4. In Tbx5(del/+) mice, a model for Holt-Oram syndrome, and Shox2(-/-) mice, we show that the T-box transcription factor Tbx5 is a regulator of Shox2 expression in the inflow tract and that Bmp4 is regulated by Shox2 in this compartment of the embryonic heart. In addition, we could show that Tbx5 acts cooperatively with Nkx2.5 to regulate the expression of Shox2 and Bmp4. This work establishes a link between Tbx5, Shox2 and Bmp4 in the pacemaker region of the developing heart and thus contributes to the unraveling of the intricate interplay between the heart-specific transcriptional machinery and developmental signaling pathways.
Figures

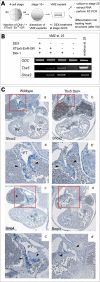
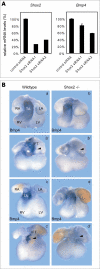

References
-
- Bruneau B.G. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. - PubMed
-
- Boyett M.R., Honjo H., Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc. Res. 2000;47:658–687. - PubMed
-
- Christoffels V.M., Smits G.J., Kispert A., Moorman A.F. Development of the pacemaker tissues of the heart. Circ. Res. 2010;106:240–254. - PubMed
-
- Blaschke R.J., Hahurij N.D., Kuijper S., Just S., Wisse L.J., Deissler K., Maxelon T., Anastassiadis K., Spitzer J., Hardt S.E., et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

