Measurement methods and accuracy in copy number variation: failure to replicate associations of beta-defensin copy number with Crohn's disease
- PMID: 20858604
- PMCID: PMC2989891
- DOI: 10.1093/hmg/ddq411
Measurement methods and accuracy in copy number variation: failure to replicate associations of beta-defensin copy number with Crohn's disease
Abstract
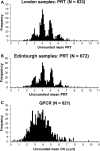
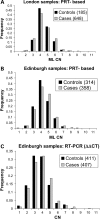
The copy number variation in beta-defensin genes on human chromosome 8 has been proposed to underlie susceptibility to inflammatory disorders, but presents considerable challenges for accurate typing on the scale required for adequately powered case-control studies. In this work, we have used accurate methods of copy number typing based on the paralogue ratio test (PRT) to assess beta-defensin copy number in more than 1500 UK DNA samples including more than 1000 cases of Crohn's disease. A subset of 625 samples was typed using both PRT-based methods and standard real-time PCR methods, from which direct comparisons highlight potentially serious shortcomings of a real-time PCR assay for typing this variant. Comparing our PRT-based results with two previous studies based only on real-time PCR, we find no evidence to support the reported association of Crohn's disease with either low or high beta-defensin copy number; furthermore, it is noteworthy that there are disagreements between different studies on the observed frequency distribution of copy number states among European controls. We suggest safeguards to be adopted in assessing and reporting the accuracy of copy number measurement, with particular emphasis on integer clustering of results, to avoid reporting of spurious associations in future case-control studies.
Figures

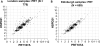

References
-
- Hollox E.J., Armour J.A.L., Barber J.C.K. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am. J. Hum. Genet. 2003;73:591–600. doi:10.1086/378157. - DOI - PMC - PubMed
-
- Hollox E.J., Davies J., Griesenbach U., Burgess J., Alton E.W.F.W., Armour J.A.L. Beta-defensin genomic copy number is not a modifier locus for cystic fibrosis. J. Negat. Results Biomed. 2005;4:9. doi:10.1186/1477-5751-4-9. - DOI - PMC - PubMed
-
- Linzmeier R.M., Ganz T. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in alpha- and beta-defensin regions at 8p22–p23. Genomics. 2005;86:423–30. doi:10.1016/j.ygeno.2005.06.003. - DOI - PubMed
-
- Hollox E.J., Barber J.C.K., Brookes A.J., Armour J.A.L. Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 2008;18:1686–1697. doi:10.1101/gr.080945.108. - DOI - PubMed
-
- Aldhous M., Noble C., Satsangi J. Dysregulation of human beta-defensin-2 protein in inflammatory bowel disease. PLoS ONE. 2009;4:e6285. doi:10.1371/journal.pone.0006285. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

