Strong and oriented immobilization of single domain antibodies from crude bacterial lysates for high-throughput compatible cost-effective antibody array generation
- PMID: 20859568
- PMCID: PMC3063850
- DOI: 10.1039/c005279e
Strong and oriented immobilization of single domain antibodies from crude bacterial lysates for high-throughput compatible cost-effective antibody array generation
Abstract
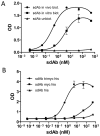
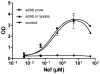
Antibody microarrays are among the novel class of rapidly emerging proteomic technologies that will allow us to efficiently perform specific diagnoses and proteomic analysis. Recombinant antibody fragments are especially suited for this approach but their stability is often a limiting factor. Camelids produce functional antibodies devoid of light chains (HCAbs) of which the single N-terminal domain is fully capable of antigen binding. When produced as an independent domain, these so-called single domain antibody fragments (sdAbs) have several advantages for biotechnological applications thanks to their unique properties of size (15 kDa), stability, solubility, and expression yield. These features should allow sdAbs to outperform other antibody formats in a number of applications, notably as capture molecules for antibody arrays. In this study, we have produced antibody microarrays using direct and oriented immobilization of sdAbs, produced in crude bacterial lysates, to generate a proof-of-principle of a high-throughput compatible array design. Several sdAb immobilization strategies have been explored. Immobilization of in vivo biotinylated sdAbs by direct spotting of bacterial lysate on streptavidin and sandwich detection was developed to achieve high sensitivity and specificity, whereas immobilization of "multi-tagged" sdAbs via anti-tag antibodies and a direct labeled sample detection strategy was optimized for the design of high-density antibody arrays for high-throughput proteomics and identification of potential biomarkers.
Figures



Similar articles
-
Single-domain antibodies: a versatile and rich source of binders for breast cancer diagnostic approaches.Mol Biosyst. 2012 Sep;8(9):2385-94. doi: 10.1039/c2mb25063b. Epub 2012 Jul 6. Mol Biosyst. 2012. PMID: 22772166
-
Comparison of single domain antibody immobilization strategies evaluated by surface plasmon resonance.J Immunol Methods. 2013 Feb 28;388(1-2):68-77. doi: 10.1016/j.jim.2012.11.014. Epub 2012 Dec 20. J Immunol Methods. 2013. PMID: 23261918
-
Phenylboronic acid polymer brush-enabled oriented and high density antibody immobilization for sensitive microarray immunoassay.Colloids Surf B Biointerfaces. 2014 Sep 1;121:21-6. doi: 10.1016/j.colsurfb.2014.05.031. Epub 2014 May 28. Colloids Surf B Biointerfaces. 2014. PMID: 24929524
-
Single Domain Antibody application in bacterial infection diagnosis and neutralization.Front Immunol. 2022 Sep 29;13:1014377. doi: 10.3389/fimmu.2022.1014377. eCollection 2022. Front Immunol. 2022. PMID: 36248787 Free PMC article. Review.
-
Enhancing Stability of Camelid and Shark Single Domain Antibodies: An Overview.Front Immunol. 2017 Jul 25;8:865. doi: 10.3389/fimmu.2017.00865. eCollection 2017. Front Immunol. 2017. PMID: 28791022 Free PMC article. Review.
Cited by
-
Hapten mediated display and pairing of recombinant antibodies accelerates assay assembly for biothreat countermeasures.Sci Rep. 2012;2:807. doi: 10.1038/srep00807. Epub 2012 Nov 12. Sci Rep. 2012. PMID: 23150778 Free PMC article.
-
A Cell-free Expression Pipeline for the Generation and Functional Characterization of Nanobodies.Front Bioeng Biotechnol. 2022 Apr 28;10:896763. doi: 10.3389/fbioe.2022.896763. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35573250 Free PMC article.
-
Nanobody-derived nanobiotechnology tool kits for diverse biomedical and biotechnology applications.Int J Nanomedicine. 2016 Jul 21;11:3287-303. doi: 10.2147/IJN.S107194. eCollection 2016. Int J Nanomedicine. 2016. PMID: 27499623 Free PMC article. Review.
-
Biotechnological applications of recombinant single-domain antibody fragments.Microb Cell Fact. 2011 Jun 9;10:44. doi: 10.1186/1475-2859-10-44. Microb Cell Fact. 2011. PMID: 21658216 Free PMC article. Review.
-
Method for Sorting and Pairwise Selection of Nanobodies for the Development of Highly Sensitive Sandwich Immunoassays.Anal Chem. 2015 Dec 1;87(23):11907-14. doi: 10.1021/acs.analchem.5b03561. Epub 2015 Nov 18. Anal Chem. 2015. PMID: 26544909 Free PMC article.
References
-
- Ekins R, Chu F, Biggart E. Multispot, multianalyte, immunoassay. Ann Biol Clin (Paris) 1990;48:655–666. - PubMed
-
- Ekins R, Chu FW. Microarrays: their origins and applications. Trends Biotechnol. 1999;17:217–218. - PubMed
-
- Marcus K, Joppich C, May C, Pfeiffer K, et al. High-resolution 2DE. Methods Mol Biol. 2009;519:221–240. - PubMed
-
- Joos T, Bachmann J. Protein microarrays: potentials and limitations. Front Biosci. 2009;14:4376–4385. - PubMed
-
- Renberg B, Nordin J, Merca A, Uhlen M, et al. Affibody molecules in protein capture microarrays: evaluation of multidomain ligands and different detection formats. J Proteome Res. 2007;6:171–179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

