Solution structure, mechanism of replication, and optimization of an unnatural base pair
- PMID: 20859962
- PMCID: PMC3332063
- DOI: 10.1002/chem.201000959
Solution structure, mechanism of replication, and optimization of an unnatural base pair
Abstract
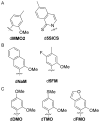
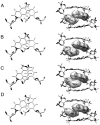
As part of an ongoing effort to expand the genetic alphabet for in vitro and eventual in vivo applications, we have synthesized a wide variety of predominantly hydrophobic unnatural base pairs and evaluated their replication in DNA. Collectively, the results have led us to propose that these base pairs, which lack stabilizing edge-on interactions, are replicated by means of a unique intercalative mechanism. Here, we report the synthesis and characterization of three novel derivatives of the nucleotide analogue dMMO2, which forms an unnatural base pair with the nucleotide analogue d5SICS. Replacing the para-methyl substituent of dMMO2 with an annulated furan ring (yielding dFMO) has a dramatically negative effect on replication, while replacing it with a methoxy (dDMO) or with a thiomethyl group (dTMO) improves replication in both steady-state assays and during PCR amplification. Thus, dTMO-d5SICS, and especially dDMO-d5SICS, represent significant progress toward the expansion of the genetic alphabet. To elucidate the structure-activity relationships governing unnatural base pair replication, we determined the solution structure of duplex DNA containing the parental dMMO2-d5SICS pair, and also used this structure to generate models of the derivative base pairs. The results strongly support the intercalative mechanism of replication, reveal a surprisingly high level of specificity that may be achieved by optimizing packing interactions, and should prove invaluable for the further optimization of the unnatural base pair.
Figures
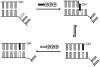

Similar articles
-
Optimization of an unnatural base pair toward natural-like replication.J Am Chem Soc. 2009 Mar 11;131(9):3246-52. doi: 10.1021/ja807853m. J Am Chem Soc. 2009. PMID: 19256568 Free PMC article.
-
Expanding the scope of replicable unnatural DNA: stepwise optimization of a predominantly hydrophobic base pair.J Am Chem Soc. 2013 Apr 10;135(14):5408-19. doi: 10.1021/ja312148q. Epub 2013 Apr 2. J Am Chem Soc. 2013. PMID: 23547847 Free PMC article.
-
Major groove substituents and polymerase recognition of a class of predominantly hydrophobic unnatural base pairs.Chemistry. 2012 Jan 23;18(4):1231-9. doi: 10.1002/chem.201102066. Epub 2011 Dec 21. Chemistry. 2012. PMID: 22190386 Free PMC article.
-
Beyond A, C, G and T: augmenting nature's alphabet.Curr Opin Chem Biol. 2003 Dec;7(6):727-33. doi: 10.1016/j.cbpa.2003.10.011. Curr Opin Chem Biol. 2003. PMID: 14644182 Review.
-
Unnatural base pair systems for sensing and diagnostic applications.Expert Rev Mol Diagn. 2011 Apr;11(3):321-31. doi: 10.1586/erm.11.5. Expert Rev Mol Diagn. 2011. PMID: 21463241 Review.
Cited by
-
The RNA World: molecular cooperation at the origins of life.Nat Rev Genet. 2015 Jan;16(1):7-17. doi: 10.1038/nrg3841. Epub 2014 Nov 11. Nat Rev Genet. 2015. PMID: 25385129 Review.
-
Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet.Proc Natl Acad Sci U S A. 2012 Jul 24;109(30):12005-10. doi: 10.1073/pnas.1205176109. Epub 2012 Jul 6. Proc Natl Acad Sci U S A. 2012. PMID: 22773812 Free PMC article.
-
Discovery, implications and initial use of semi-synthetic organisms with an expanded genetic alphabet/code.Philos Trans R Soc Lond B Biol Sci. 2023 Feb 27;378(1871):20220030. doi: 10.1098/rstb.2022.0030. Epub 2023 Jan 11. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 36633274 Free PMC article. Review.
-
The strength of the template effect attracting nucleotides to naked DNA.Nucleic Acids Res. 2014 Jun;42(11):7409-20. doi: 10.1093/nar/gku314. Epub 2014 May 29. Nucleic Acids Res. 2014. PMID: 24875480 Free PMC article.
-
Method for Rapid Analysis of Mutant RNA Polymerase Activity on Templates Containing Unnatural Nucleotides.Int J Mol Sci. 2021 May 14;22(10):5186. doi: 10.3390/ijms22105186. Int J Mol Sci. 2021. PMID: 34069057 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

