The transcriptional repressor ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression
- PMID: 20861012
- PMCID: PMC2998095
- DOI: 10.1074/jbc.M110.175182
The transcriptional repressor ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression
Abstract
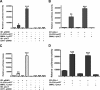
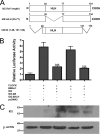
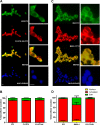
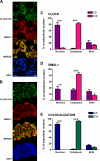
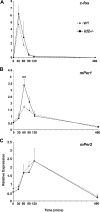
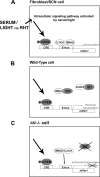
ID2 is a rhythmically expressed HLH transcriptional repressor. Deletion of Id2 in mice results in circadian phenotypes, highlighted by disrupted locomotor activity rhythms and an enhanced photoentrainment response. ID2 can suppress the transactivation potential of the positive elements of the clock, CLOCK-BMAL1, on mPer1 and clock-controlled gene (CCG) activity. Misregulation of CCGs is observed in Id2(-/-) liver, and mutant mice exhibit associated alterations in lipid homeostasis. These data suggest that ID2 contributes to both input and output components of the clock and that this may be via interaction with the bHLH clock proteins CLOCK and BMAL1. The aim of the present study was to explore this potential interaction. Coimmunoprecipitation analysis revealed the capability of ID2 to complex with both CLOCK and BMAL1, and mammalian two-hybrid analysis revealed direct interactions of ID2, ID1 and ID3 with CLOCK and BMAL1. Deletion of the ID2 HLH domain rendered ID2 ineffective at inhibiting CLOCK-BMAL1 transactivation, suggesting that interaction between the proteins is via the HLH region. Immunofluorescence analysis revealed overlapping localization of ID2 with CLOCK and BMAL1 in the cytoplasm. Overexpression of CLOCK and BMAL1 in the presence of ID2 resulted in a significant reduction in their nuclear localization, revealing that ID2 can sequester CLOCK and BMAL1 to the cytoplasm. Serum stimulation of Id2(-/-) mouse embryonic fibroblasts resulted in an enhanced induction of mPer1 expression. These data provide the basis for a molecular mechanism through which ID2 could regulate aspects of both clock input and output through a time-of-day specific interaction with CLOCK and BMAL1.
Figures

References
-
- Reppert S. M., Weaver D. R. (2001) Annu. Rev. Physiol. 63, 647–676 - PubMed
-
- Dunlap J. C., Loros J. J., DeCoursey P. J. (2004) Chronobiology: Biological Timekeeping, Sinauer Associates, Sunderland, MA
-
- Peirson S. N., Butler J. N., Duffield G. E., Takher S., Sharma P., Foster R. G. (2006) Biochem. Biophys. Res. Commun. 351, 800–807 - PubMed
-
- Balsalobre A., Damiola F., Schibler U. (1998) Cell 93, 929–937 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

