Paradoxical instability-activity relationship defines a novel regulatory pathway for retinoblastoma proteins
- PMID: 20861300
- PMCID: PMC2982090
- DOI: 10.1091/mbc.E10-06-0520
Paradoxical instability-activity relationship defines a novel regulatory pathway for retinoblastoma proteins
Abstract
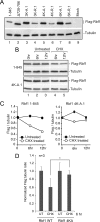
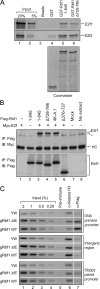
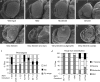
The retinoblastoma (RB) transcriptional corepressor and related family of pocket proteins play central roles in cell cycle control and development, and the regulatory networks governed by these factors are frequently inactivated during tumorigenesis. During normal growth, these proteins are subject to tight control through at least two mechanisms. First, during cell cycle progression, repressor potential is down-regulated by Cdk-dependent phosphorylation, resulting in repressor dissociation from E2F family transcription factors. Second, RB proteins are subject to proteasome-mediated destruction during development. To better understand the mechanism for RB family protein instability, we characterized Rbf1 turnover in Drosophila and the protein motifs required for its destabilization. We show that specific point mutations in a conserved C-terminal instability element strongly stabilize Rbf1, but strikingly, these mutations also cripple repression activity. Rbf1 is destabilized specifically in actively proliferating tissues of the larva, indicating that controlled degradation of Rbf1 is linked to developmental signals. The positive linkage between Rbf1 activity and its destruction indicates that repressor function is governed in a manner similar to that described by the degron theory of transcriptional activation. Analogous mutations in the mammalian RB family member p107 similarly induce abnormal accumulation, indicating substantial conservation of this regulatory pathway.
Figures




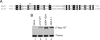
References
-
- Binne U. K., Classon M. K., Dick F. A., Wei W., Rape M., Kaelin W. G., Jr, Naar A. M., Dyson N. J. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell. Biol. 2007;9:225–232. - PubMed
-
- Boyer S. N., Wazer D. E., Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. - PubMed
-
- Brand A. H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Classon M., Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer. 2002;2:910–917. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

