Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species
- PMID: 20861340
- PMCID: PMC3020881
- DOI: 10.1128/JCM.01191-10
Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species
Abstract
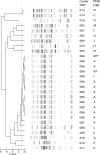
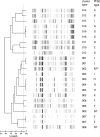
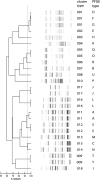
Many bacterial typing methods are specific for one species only, time-consuming, or poorly reproducible. DiversiLab (DL; bioMérieux) potentially overcomes these limitations. In this study, we evaluated the DL system for the identification of hospital outbreaks of a number bacterial species. Appropriately typed clinical isolates were tested with DL. DL typing agreed with pulsed-field gel electrophoresis (PFGE) for Acinetobacter (n = 26) and Stenotrophomonas maltophilia (n = 13) isolates. With two exceptions, DL typing of Klebsiella isolates (n = 23) also correlated with PFGE, and in addition, PFGE-nontypeable (PFGE-NT) isolates could be typed. Enterobacter (n = 28) results also correlated with PFGE results; also, PFGE-NT isolates could be clustered. In a larger study (n = 270), a cluster of 30 isolates was observed that could be subdivided by PFGE. The results for Escherichia coli (n = 38) correlated less well with an experimental multilocus variable number of tandem repeats analysis (MLVA) scheme. Pseudomonas aeruginosa (n = 52) showed only a limited number of amplification products for most isolates. When multiple Pseudomonas isolates were assigned to a single type in DL, all except one showed multiple multilocus sequence types. Methicillin-resistant Staphylococcus aureus generally also showed a limited number of amplification products. Isolates that belonged to different outbreaks by other typing methods, including PFGE, spa typing, and MLVA, were grouped together in a number of cases. For Enterococcus faecium, the limited variability of the amplification products obtained made interpretation difficult and correlation with MLVA and esp gene typing was poor. All of the results are reflected in Simpson's index of diversity and adjusted Rand's and Wallace's coefficients. DL is a useful tool to help identify hospital outbreaks of Acinetobacter spp., S. maltophilia, the Enterobacter cloacae complex, Klebsiella spp., and, to a somewhat lesser extent, E. coli. In our study, DL was inadequate for P. aeruginosa, E. faecium, and MRSA. However, it should be noted that for the identification of outbreaks, epidemiological data should be combined with typing results.
Figures



References
-
- Bonacorsi, S., P. Bidet, F. Mahjoub, P. Mariani-Kurkdjian, S. Ait-Ifrane, C. Courroux, and E. Bingen. 2009. Semi-automated rep-PCR for rapid differentiation of major clonal groups of Escherichia coli meningitis strains. Int. J. Med. Microbiol. 299:402-409. - PubMed
-
- Carretto, E., D. Barbarini, C. Farina, A. Grosini, P. Nicoletti, E. Manso, and the APSI Acinetobacter Study Group, Italy. 2008. Use of the DiversiLab semiautomated repetitive-sequence-based polymerase chain reaction for epidemiologic analysis on Acinetobacter baumannii isolates in different Italian hospitals. Diagn. Microbiol. Infect. Dis. 60:1-7. - PubMed
-
- de Bruijn, F. J., J. R. Lupski, and G. M. Weinstock. 1998. Bacterial genomes: physical structure and analysis. Chapman & Hall, New York, NY.
-
- Doléans-Jordheim, A., B. Cournoyer, E. Bergeron, J. Croizé, H. Salord, J. André, M. A. Mazoyer, F. N. Renaud, and J. Freney. 2009. Reliability of Pseudomonas aeruginosa semi-automated rep-PCR genotyping in various epidemiological situations. Eur. J. Clin. Microbiol. Infect. Dis. 28:1105-1111. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

