Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing
- PMID: 20861475
- PMCID: PMC3053293
- DOI: 10.1167/iovs.10-6180
Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing
Abstract
Purpose: To determine whether massively parallel next-generation DNA sequencing offers rapid and efficient detection of disease-causing mutations in patients with monogenic inherited diseases. Retinitis pigmentosa (RP) is a challenging application for this technology because it is a monogenic disease in individuals and families but is highly heterogeneous in patient populations. RP has multiple patterns of inheritance, with mutations in many genes for each inheritance pattern and numerous, distinct, disease-causing mutations at each locus; further, many RP genes have not been identified yet.
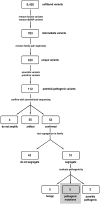
Methods: Next-generation sequencing was used to identify mutations in pairs of affected individuals from 21 families with autosomal dominant RP, selected from a cohort of families without mutations in "common" RP genes. One thousand amplicons targeting 249,267 unique bases of 46 candidate genes were sequenced with the 454GS FLX Titanium (Roche Diagnostics, Indianapolis, IN) and GAIIx (Illumina/Solexa, San Diego, CA) platforms.
Results: An average sequence depth of 70× and 125× was obtained for the 454GS FLX and GAIIx platforms, respectively. More than 9000 sequence variants were identified and analyzed, to assess the likelihood of pathogenicity. One hundred twelve of these were selected as likely candidates and tested for segregation with traditional di-deoxy capillary electrophoresis sequencing of additional family members and control subjects. Five disease-causing mutations (24%) were identified in the 21 families.
Conclusion: This project demonstrates that next-generation sequencing is an effective approach for detecting novel, rare mutations causing heterogeneous monogenic disorders such as RP. With the addition of this technology, disease-causing mutations can now be identified in 65% of autosomal dominant RP cases.
Figures


References
-
- Albert TJ, Molla MN, Muzny DM, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905 - PubMed
-
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909 - PubMed
-
- Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

