Clonal analysis of the microbiota of severe early childhood caries
- PMID: 20861633
- PMCID: PMC2975730
- DOI: 10.1159/000320158
Clonal analysis of the microbiota of severe early childhood caries
Abstract
Background/aims: Severe early childhood caries is a microbial infection that severely compromises the dentition of young children. The aim of this study was to characterize the microbiota of severe early childhood caries.
Methods: Dental plaque samples from 2- to 6-year-old children were analyzed using 16S rRNA gene cloning and sequencing, and by specific PCR amplification for Streptococcus mutans and Bifidobacteriaceae species.
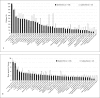
Results: Children with severe caries (n = 39) had more dental plaque and gingival inflammation than caries-free children (n = 41). Analysis of phylotypes from operational taxonomic unit analysis of 16S rRNA clonal metalibraries from severe caries and caries-free children indicated that while libraries differed significantly (p < 0.0001), there was increased diversity than detected in this clonal analysis. Using the Human Oral Microbiome Database, 139 different taxa were identified. Within the limits of this study, caries-associated taxa included Granulicatella elegans (p < 0.01) and Veillonella sp. HOT-780 (p < 0.01). The species associated with caries-free children included Capnocytophaga gingivalis (p < 0.01), Abiotrophia defectiva (p < 0.01), Lachnospiraceae sp. HOT-100 (p < 0.05), Streptococcus sanguinis (p < 0.05) and Streptococcus cristatus (p < 0.05). By specific PCR, S. mutans (p < 0.005) and Bifidobacteriaceae spp. (p < 0.0001) were significantly associated with severe caries.
Conclusion: Clonal analysis of 80 children identified a diverse microbiota that differed between severe caries and caries-free children, but the association of S. mutans with caries was from specific PCR analysis, not from clonal analysis, of samples.
Copyright © 2010 S. Karger AG, Basel.
Figures




References
-
- Beltran-Aguilar ED, Barker LK, Canto MT, Dye BA, Gooch BF, Griffin SO, Hyman J, Jaramillo F, Kingman A, Nowjack-Raymer R, Selwitz RH, Wu T. Surveillance for dental caries, dental sealants, tooth retention, edentulism, and enamel fluorosis – United States, 1988–1994 and 1999–2002. MMWR Surveill Summ. 2005;54:1–43. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series BMethodol. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

