Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing
- PMID: 20862214
- PMCID: PMC2942828
- DOI: 10.1371/journal.pone.0012745
Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing
Abstract
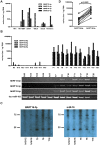
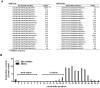
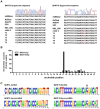
Virus-encoded microRNAs (miRNAs) have been shown to regulate a variety of biological processes involved in viral infection and viral-associated pathogenesis. Epstein-Barr virus (EBV) is a herpesvirus implicated in nasopharyngeal carcinoma (NPC) and other human malignancies. EBV-encoded miRNAs were among the first group of viral miRNAs identified. To understand the roles of EBV miRNAs in the pathogenesis of NPC, we utilized deep sequencing technology to characterize the EBV miRNA transcriptome in clinical NPC tissues. We obtained more than 110,000 sequence reads in NPC samples and identified 44 EBV BART miRNAs, including four new mature miRNAs derived from previously identified BART miRNA precursor hairpins. Further analysis revealed extensive sequence variations (isomiRs) of EBV miRNAs, including terminal isomiRs at both the 5' and 3' ends and nucleotide variants. Analysis of EBV genomic sequences indicated that the majority of EBV miRNA nucleotide variants resulted from post-transcriptional modifications. Read counts of individual EBV miRNA in NPC tissue spanned from a few reads to approximately 18,000 reads, confirming the wide expression range of EBV miRNAs. Several EBV miRNAs were expressed at levels similar to highly abundant human miRNAs. Sequence analysis revealed that most of the highly abundant EBV miRNAs share their seed sequences (nucleotides 2-7) with human miRNAs, suggesting that seed sequence content may be an important factor underlying the differential accumulation of BART miRNAs. Interestingly, many of these human miRNAs have been found to be dysregulated in human malignancies, including NPC. These observations not only provide a potential linkage between EBV miRNAs and human malignancy but also suggest a highly coordinated mechanism through which EBV miRNAs may mimic or compete with human miRNAs to affect cellular functions.
Conflict of interest statement
Figures
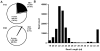


References
-
- Burgos JS. Involvement of the Epstein-Barr virus in the nasopharyngeal carcinoma pathogenesis. Med Oncol. 2005;22:113–121. - PubMed
-
- Middeldorp JM, Pegtel DM. Multiple roles of LMP1 in Epstein-Barr virus induced immune escape. Semin Cancer Biol. 2008;18:388–396. - PubMed
-
- Iwakiri D, Takada K. Role of EBERs in the pathogenesis of EBV infection. Adv Cancer Res. 2010;107:119–136. - PubMed
-
- Yoshizaki T. Promotion of metastasis in nasopharyngeal carcinoma by Epstein-Barr virus latent membrane protein-1. Histol Histopathol. 2002;17:845–850. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

