Petri Nets with Fuzzy Logic (PNFL): reverse engineering and parametrization
- PMID: 20862218
- PMCID: PMC2942832
- DOI: 10.1371/journal.pone.0012807
Petri Nets with Fuzzy Logic (PNFL): reverse engineering and parametrization
Abstract
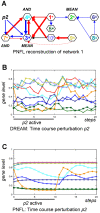
Background: The recent DREAM4 blind assessment provided a particularly realistic and challenging setting for network reverse engineering methods. The in silico part of DREAM4 solicited the inference of cycle-rich gene regulatory networks from heterogeneous, noisy expression data including time courses as well as knockout, knockdown and multifactorial perturbations.
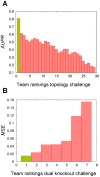
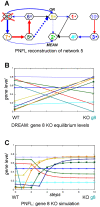
Methodology and principal findings: We inferred and parametrized simulation models based on Petri Nets with Fuzzy Logic (PNFL). This completely automated approach correctly reconstructed networks with cycles as well as oscillating network motifs. PNFL was evaluated as the best performer on DREAM4 in silico networks of size 10 with an area under the precision-recall curve (AUPR) of 81%. Besides topology, we inferred a range of additional mechanistic details with good reliability, e.g. distinguishing activation from inhibition as well as dependent from independent regulation. Our models also performed well on new experimental conditions such as double knockout mutations that were not included in the provided datasets.
Conclusions: The inference of biological networks substantially benefits from methods that are expressive enough to deal with diverse datasets in a unified way. At the same time, overly complex approaches could generate multiple different models that explain the data equally well. PNFL appears to strike the balance between expressive power and complexity. This also applies to the intuitive representation of PNFL models combining a straightforward graphical notation with colloquial fuzzy parameters.
Conflict of interest statement
Figures





References
-
- de Jong H. Modeling and simulation of genetic regulatory systems: a literature review. J Comput Biol. 2002;9(1):67–103. - PubMed
-
- Brazhnik P, de la Fuente A, Mendes P. Gene networks: how to put the function in genomics. Trends Biotechnol. 2002;20(11):467–72. - PubMed
-
- Li H, Xuan J, Wang Y, Zhan M. Inferring regulatory networks. Front Biosci. 2008;13:263–75. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

